Abstract
Cytotoxin-associated gene A (CagA) diversity with regard to EPIYA-A, -B, -C, or -D phosphorylation motifs may play an important role in Helicobacter pylori pathogenesis, and therefore determination of these motifs in H. pylori clinical isolates can become a useful prognostic tool. We propose a strategy for the accurate determination of CagA EPIYA motifs in clinical strains, based upon one-step PCR amplification using primers that flank the EPIYA coding region. We thus analyzed 135 H. pylori isolates derived from 75 adults and 60 children Greek patients. A total of 34 cases were found to be EPIYA PCR negative and were consequently verified as cagA negative by cagA-specific PCR, empty-site cagA PCR, and Western blotting. Sequencing of the remaining 101 PCR-positive amplicons confirmed that an accurate prediction of the number of EPIYA motifs on the basis of size distribution of the PCR products was feasible in all cases. Furthermore, our assay could identify closely related H. pylori subclones within the same patient, harboring different numbers of EPIYA repeats. The prevalence of CagA proteins with three EPIYA motifs (ABC) or four EPIYA motifs (ABCC) was the same within the adult and children groups. However, CagA species with more than four EPIYA motifs were observed exclusively within adults (8.6%), suggesting that CagA-positive strains may acquire additional EPIYA-C motifs throughout adulthood. Our strategy requires no initial cagA screening of the clinical isolates and can accurately predict the number of EPIYA repeats in single or multiple closely related subclones bearing different numbers of EPIYA motifs in their CagA, which may coexist within the same patient.
CagA, a 120- to 145-kDa bacterial protein, is recognized as a major etiologic determinant of Helicobacter pylori-associated gastric disease found to increase the risk for peptic ulceration (14, 15, 23), atrophic gastritis (20) and non-cardia gastric adenocarcinoma (12, 25). After H. pylori binding to the gastric epithelium, CagA has been shown to translocate into the gastric epithelial cell cytoplasm via the H. pylori type IV secretion system (24). Once injected into the epithelial cells, CagA localizes to the plasma membrane (3) and undergoes tyrosine phosphorylation (27) by multiple members of the Src family of kinases (30, 28) on specific tyrosine residues within repeating Glu-Pro-Ile-Tyr-Ala (EPIYA) motifs (9, 17). These EPIYA motifs are defined as EPIYA-A, -B -C, and -D, according to the amino acid sequence that surrounds the EPIYA sequence (17). Earlier studies have showed that CagA protein species nearly always contain EPIYA-A and EPIYA-B sites, followed by one to three repeats of EPIYA-C in Western-type H. pylori isolates (26) or EPIYA-D site in East Asian-type isolates (8, 35, 36). Thus, CagA species usually vary on the number of EPIYA-C or -D repeats in the carboxyl terminus of the protein. Phosphorylated CagA has been reported to interact with and deregulate the activity of a number of intracellular effectors relating to the hepatocyte growth factor signaling pathway, such as Src homology 2-containing protein tyrosine phosphatase 2 (16), growth factor receptor bound 2 (21), carboxyl-terminal Src kinase (29), and hepatocyte growth factor receptor/cMet (13). More specifically, tyrosine-phosphorylated CagA seems to bind and deregulate the activity of Src homology 2-containing protein tyrosine phosphatase 2 via the Western CagA-specific EPIYA-C or East Asian CagA-specific EPIYA-D site and of carboxyl-terminal Src kinase via the EPIYA-A or EPIYA-B site (22). In parallel, CagA EPIYA motifs have been suggested to play an essential role for the tethering of CagA to the membrane in a phosphorylation-independent manner (18). Consequently, CagA variability with reference to EPIYA motifs may play an important role in H. pylori pathogenesis. CagA-positive clinical strains with an increased number of EPIYA phosphorylation motifs isolated from Eastern populations have been associated with more severe active chronic gastritis and atrophy (8). An increasing number of EPIYA motifs within the Western-type CagA have been related with higher interleukin-8 secretion and more pronounced cellular elongation (6). Therefore, EPIYA motif diversity may prove useful in the prediction of H. pylori pathogenic activity, and the accurate determination of the type and number of EPIYA motifs in clinical H. pylori isolates can become a useful prognostic tool.
In a clinical microbiology laboratory, H. pylori isolation from gastric biopsies is often made by the sweeping method instead of individual colony selection and, therefore, the presence of multiple closely related subclones within an H. pylori isolate is likely to occur (10, 11). Such H. pylori subclones harboring different numbers of CagA EPIYA motifs have been observed to coexist within the same patient (5). An elegant PCR-based assay for the determination of CagA EPIYA motifs, utilizing three separate sets of primers specific for each EPIYA motif, has already been proposed (7). However, it assumes clonal uniformity with reference to EPIYA motif diversity within an isolate. Hence, during routine screening of H. pylori clinical isolates, the use of such a method for the determination of EPIYA motifs may produce ambiguous results due to the presence of closely related subclones with different numbers of CagA EPIYA motifs.
In the present study, we designed specific primers and successfully amplified the variable 3′ end of cagA gene and then sequenced the PCR products in more than 100 H. pylori clinical strains. Based on our sequencing results, the numbers and types of EPIYA motifs within CagA protein can safely be predicted by the size of the one-step PCR amplicon in more than 90% of the cases. We also confirmed that our method could accurately predict cagA presence in the isolated strains, by cagA-specific PCR, empty-site cagA PCR, and Western blot analysis of CagA expression, thus eliminating the need for initial screening of cagA status after strain isolation. Furthermore, our strategy enabled us to detect within the same patient the presence of multiple closely related infecting H. pylori subclones with different numbers of EPIYA motifs in CagA and facilitated their isolation. In this way, we determined EPIYA diversity within the CagA protein for more than 100 cagA-positive clinical H. pylori isolates from adults and children.
MATERIALS AND METHODS
Clinical isolates.
H. pylori clinical isolates (n = 135), derived from 75 adult (48 male, 27 female, mean age of 52.1 ± 1.6 years) and 60 child patients (24 male, 36 female, mean age of 9.9 ± 0.6 years) were isolated from gastric biopsies obtained during upper gastroduodenal endoscopy at the Gastroenterology Clinics of Alexandras General Hospital, Evangelismos General Hospital, and Aghia Sophia Children's Hospital. All patients were Greek in origin and signed a consent form for participation in the study. Each clinical isolate was passed twice on Chalgren's-Wilkins agar plates containing antibiotics (vancomycin [10 μg/ml], trimethoprim [10 μg/ml], polymyxin B [104 IU/liter], amphotericin B [2 μg/ml], nalidixic acid [10 μg/ml], bacitracin [30 μg/ml], fluorocytosine [5 μg/ml]; Sigma, St. Louis, MO) supplemented with 7% (vol/vol) horse blood and 1% (vol/vol) Vitox (Oxoid, Basingstoke, United Kingdom). Cultures were incubated at 37°C under microaerophilic conditions (CampyPak-Plus; Becton Dickinson, Cockeysville, MD). The total bacterial genomic DNA was extracted by using the DNeasy isolation kit (QIAGEN AS, Oslo, Norway), and the ratio of the optical density at 260 nm to that at 280 nm was greater than 1.800. H. pylori strain SS1 was also included in the study as a control strain since it has a published cagA gene sequence. RAPD [random(ly) amplified polymorphic DNA] PCR profiles of isolates were obtained utilizing the primers 1281 and D11344 as described previously (2).
Amplification and sequencing of the EPIYA-containing region of CagA.
After initial alignment of full CagA protein sequences found in GenBank, we identified conserved peptide sequences 845′-VKNGVNGTLVGN-856′ and 1011′-LNQAVSEAK-1019′, which flank the region harboring the EPIYA motifs (positions according to the H. pylori 26695 genome [32]). Based upon these peptides we designed the primers cagA2530S (5′-GTTAARAATRGTGTRAAYGG-3′, where R = A or G and Y = T or C) and cagA3000AS (5′-TTTAGCTTCTGATACCGC-3′; positions 582453 to 582977 with reference to the H. pylori 26695 genome). Screening of the oligonucleotides by BLAST analysis identified exclusively H. pylori cagA sequences (data not shown).
PCR amplification was carried out within 50-μl reaction mixtures containing 10 mM Tris, 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin, 1 U of Taq (Fermentas UAB, Vilnius, Lithuania), 200 μM concentrations of deoxynucleoside triphosphates, 0.5 μM concentrations of the primers, and 50 ng of microbial genomic DNA. The PCR conditions included an initial denaturation step at 94°C for 5 min, followed by 35 cycles at 94°C for 30 s, 50°C for 45 s, and 72°C for 45 s, and then an extension at 72°C for 5 min. The conditions of PCR amplification were optimized by using an Eppendorf Mastercycler gradient apparatus (Eppendorf AG, Hamburg, Germany) with annealing temperatures ranging from 40.0 to 60.4°C (data not shown). Single PCR products ranging from 370 to 670 bp (±25 bp) were initially visualized by agarose gel electrophoresis. They were subsequently isolated by using a QIAquick PCR purification kit (QIAGEN) and sequenced by automated sequencing on a Li-Cor DNA sequencer long read IR2 4200 (IMBB facility, Crete), utilizing IRD700-labeled cagA3000AS primer and a SequiTherm EXCEL II DNA sequencing kit-LC (Epicenter Biotechnologies, Madison, WI). The deduced peptide sequences containing the EPIYA motifs were aligned by CLUSTAL W (European Bioinformatics Institute [http://www.ebi.ac.uk/clustalw/]). During the initial developmental stages of the assay, we used the PCR product of H. pylori SS1 strain (accession number AAF63759) as a size control of a cagA nucleotide sequence coding for three EPIYA motifs. All strains were also screened for the presence of the cagA gene by PCR as described earlier (33). EPIYA PCR-negative strains were confirmed as being cagA negative by the empty site-positive PCR assay for the characterization of cagA-negative strains (1).
During initial development stages, we validated the ability of our PCR procedure to amplify the EPIYA-coding region in 61 cagA-positive samples by utilizing the primers cag2 and cag4 (26), which amplify the variable 3′ region of the cagA gene between positions 582471 and 583025 with reference to the H. pylori 26695 genome. Finally, based on our sequence data we evaluated the ability of a PCR-based assay reported earlier (7) to accurately predict the number and type of EPIYA motifs.
Analysis of CagA protein expression by Western blot analysis.
Human gastric adenocarcinoma epithelial AGS cells, cultured in F-12 Kaighn's medium (Gibco/Invitrogen, Ltd., Paisley, United Kingdom) containing 10% fetal bovine serum (Gibco) were infected with H. pylori clinical strains at a multiplicity of infection of 100 and incubated under a 5% CO2 atmosphere for 24 h. Total protein lysates from infected cells or H. pylori bacterial preparations were obtained in ice-cold lysis buffer (150 mM NaCl, 10 mM Tris-HCl [pH 7.2], 0.1% sodium dodecyl sulfate, 1% Triton X-100, 1% deoxycholate, 5 mM EDTA, 2 mM l-dithiothreitol) containing protease and phosphatase inhibitor cocktails. Lysates were centrifuged at 14,000 × g for 30 min at 4°C, and the supernatants were kept at −20°C until use. The total protein was determined by using a Pierce MicroBCA protein assay (Pierce Biotechnology, Inc., Rockford, IL). Equal protein amounts of cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (7.5% polyacrylamide) and transferred onto polyvinylidene difluoride (Immobilon-P; Millipore Corp., Bedford, MA) membranes. We performed standard Western blotting with primary anti-CagA monoclonal antibody (Austral Biologicals, San Ramon, CA) at a dilution of 1:1,000 and secondary horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G polyclonal antibody (Jackson Immunoresearch Europe, Ltd., Soham, Cambridgeshire, United Kingdom). CagA expression was detected by utilizing a chemiluminescence detection system (ECL Plus) according to the manufacturer's instructions (Amersham/GE Healthcare UK, Ltd., Buckinghamshire, United Kingdom).
Accession numbers.
All partial nucleotide cagA sequences generated in the present study have been submitted to the GenBank/EMBL/DDBJ databases (accession numbers AM279288 to AM279335, AM292553 to AM292599, and AM295786 to AM295791).
RESULTS
PCR amplification of the variable 3′ region of the cagA gene coding for EPIYA motifs.
We successfully amplified the variable 3′ region of the cagA gene in 101 of 135 H. pylori isolates by utilizing our EPIYA PCR method and observed a single-band PCR product (Fig. 1) in 91 cases and a double-band product (Fig. 2A) in 10 cases. The PCR amplicons ranged in size between 370 and 670 bp (±25 bp) and were arranged equidistantly (approximately 100 bp) in a ladder-like arrangement indicative of the presence of multiple repeated sequences. All strains negative for EPIYA PCR (n = 34) were further verified as true cagA-negative strains utilizing a cagA-specific PCR, empty-site-positive PCR assay for cagA-negative strains and Western blotting. More specifically, using cagA-specific PCR we identified only 15 cagA-negative strains by the absence of the characteristic 180-bp PCR product (data not shown). However, utilizing the empty-site-positive PCR assay we identified as true cagA negative all 34 strains that were EPIYA PCR negative. Finally, no expression of CagA protein was detected by Western blotting in whole-cell bacterial lysates from all EPIYA PCR-negative strains (data not shown). The remaining 101 strains that gave a positive EPIYA PCR product were all confirmed as cagA positive by cagA-specific PCR and Western blotting of whole-cell bacterial lysates (data not shown). Of these strains, 43 were isolated from children (21 males, 22 females) and 58 were from adult patients (39 male, 19 female). No significant difference in EPIYA PCR positivity was detected between the adult and children populations or between genders. Finally, in 61 strains we also utilized the primers cag2 and cag4 (26), which amplify the EPIYA-coding region in the cagA gene between positions 582471 and 583025 with reference to the H. pylori 26695 genome. Although three cases were not amplified by the cag2 and cag4 primers, we observed no significant difference (P = 0.244 [Fisher's exact test]) in the ability of the two PCR methods to amplify the EPIYA-coding region (data not shown).
FIG. 1.
Electrophoretic analysis of EPIYA PCR products. The DNA from H. pylori clinical strains (lanes 1 to 8) was amplified by EPIYA PCR, and the PCR products were analyzed on a 1.5% agarose gel. The samples in lanes 9 to 11 are sequenced EPIYA PCR products from cagA-positive strains with ABCC (lane 9; Hp63, AM292557), ABC (lane 10; Hp5, AM292581), and AB (lane 11; Hp51, AM292592) combinations. Note the ladder-like distribution in the molecular weights of the samples, which is indicative of the presence of multiple repeats. Based on their size, the EPIYA number predictions were 3 (samples 3, 4, 5, and 7), 4 (samples 1 and 8), and 5 (sample 2). Sample 6 was a cagA-negative isolate.
FIG. 2.
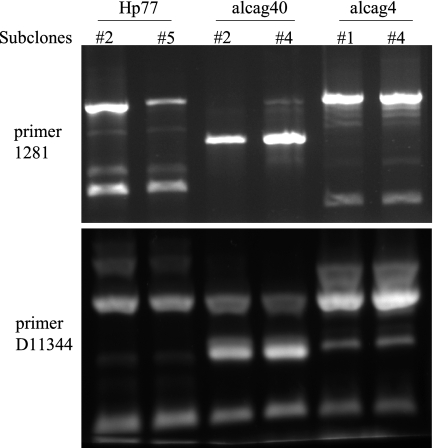
H. pylori closely related paired subclones Hp77-2-Hp77-5, alcag4-1-alcag4-4, and alcag40-2-alcag40-4 isolated from the same respective patients, differing in the number of repeating EPIYA-C motifs. (A) Agarose gel mobilities of EPIYA PCR amplicons in the mixed isolates (lanes 1, 4, and 7) and the respective isolated paired subclones (lanes 2 and 3, 5 and 6, and 8 and 9). The sizes of the PCR amplicons specific for two (Hp51, AM292592), three (Hp5, AM292581), four (Hp63, AM292557), and five (alcag202, AM279288) EPIYA motifs are depicted in lane 10. (B) Aligned nucleotide sequences of the EPIYA PCR amplicons for the paired subclones. (C) Aligned deduced peptide sequences of CagA protein with different numbers of EPIYA-C repeats in the paired subclones. (D) Functional expression of CagA protein from the paired subclones, following in vitro infection of adenocarcinoma cell line AGS. Note the presence of double bands in the mixed clinical isolates (lanes 1, 4, and 7) and the corresponding single bands in the individual subclones (lanes 2 and 3, 5 and 6, and 8 and 9). The GenBank/EMBL/DDBJ accession numbers are as follows: Hp77-2, AM292594; Hp77-5, AM292593; alcag4-1, AM292554; alcag4-4, AM292555; alcag40-2, AM292577; and alcag40-4, AM292578.
EPIYA PCR analysis of the H. pylori SS1 strain yielded an amplification product of 470 ± 25 bp, and this was used as a size control for sequences encoding for three EPIYA motifs. Thus, initial prediction on the expected number of EPIYA repeats was possible by direct comparison of the corresponding sizes of PCR amplicons.
In 10 isolates following EPIYA PCR, we detected the presence of a double band with sizes corresponding to different numbers of EPIYA repeats (Fig. 2A). We verified by isolation and sequencing that those were cagA-specific sequences, an indication of the presence of at least two infecting strains, within the same patient. We successfully separated those subclones by limiting dilution, H. pylori colony selection, and screening of individual colonies for a single PCR amplicon by our EPIYA PCR (Fig. 2A). We utilized RAPD PCR to assess the clonal relatedness of the isolated subclones and verified that these strains were very closely related to each other (Fig. 3). More specifically, RAPD profiles with primer D11344 were identical between subclones, and those obtained by primer 1281 showed very close relatedness. Collectively these data suggest that our EPIYA PCR assay can (i) accurately predict the presence of the cagA gene in H. pylori clinical isolates, (ii) efficiently amplify the variable cagA gene 3′-region encoding for the EPIYA motifs, and (iii) detect the presence and facilitate the isolation of the individual infecting subclones.
FIG. 3.
RAPD profiles of closely related paired subclones Hp77-2-Hp77-5, alcag4-2-alcag4-4, and alcag40-2-alcag40-4 isolated from the same respective patients. Primers 1281 and D11344 were used as described previously (2). The patterns indicate high relatedness among the paired subclones.
CagA diversity with respect to the number and type of EPIYA motifs.
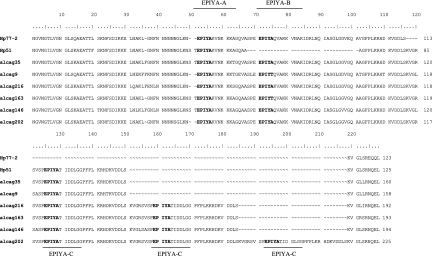
All amplified EPIYA PCR products were sequenced, and the deduced peptide sequences were aligned by using CLUSTAL W (Fig. 4). The H. pylori SS1 strain PCR product when sequenced was found to be identical to the already published sequence (accession number AAF63759; and data not shown). Upon alignment of the deduced protein sequences in our clinical samples, we observed three types of EPIYA motifs, namely, (i) EPIYA-A, EPIYAKVNKKK(A/T/V/S)GQ; EPIYA-B, EPIY(A/T)(Q/K)VAKKVNAKI; and EPIYA-C, EPIYATIDDLG (Fig. 4). We found no strains within our population harboring the Eastern type of EPIYA-D (EPIYATIDFDEANQAG). Our initial predictions about the number of EPIYA repeats, based upon the size of the PCR amplicons, were all verified on the basis of the nucleotide and the deduced peptide sequences. Furthermore, in all cases where we isolated from the same patient two closely related subclones predicted to have different number of EPIYA motifs, the amplified nucleotide sequence and the subsequent aligned peptide sequences obtained verified our original prediction (Fig. 2B and C). Upon comparison, sequences were found to be identical on a nucleotide basis, outside the 102-bp sequence repeats coding for the 34-amino-acid peptide segment containing the additional EPIYA-C motifs (Fig. 2B and C). In these closely related subclones, we further verified the expression of the CagA proteins with different numbers of EPIYA motifs by Western blot analysis of total protein lysates after infection of AGS gastric adenocarcinoma epithelial cells with the mixed isolates, as well as the individual isolated subclones (Fig. 2D). These data suggest that our EPIYA PCR can effectively identify the number of EPIYA motifs within the CagA protein in H. pylori isolates both in single and in multiple closely related subclones bearing different numbers of EPIYA motifs in their CagA proteins.
FIG. 4.
Alignment of partial CagA peptide sequences derived from our analysis, depicting a representative collection of diverse CagA species with respect to the number and types of EPIYA motifs. The GenBank/EMBL/DDBJ accession numbers are as follows: Hp77-2, AM292594; Hp51, AM292592; alcag35, AM279305; alcag9, AM279304; alcag216, AM279293; alcag163, AM279292; alcag146, AM279291; and alcag202, AM279288.
Comparison of PCR methods for determination of the number of EPIYA motifs.
An elegant PCR method utilizing three different sets of primers specific for each EPIYA type coding sequence had been proposed for the determination of the number of EPIYA motifs (7). Having obtained the sequences for the variable 3′ region of cagA gene, we compared that PCR method and our EPIYA PCR with regard to their ability to predict the correct number of EPIYA motifs present in 31 single-clone H. pylori isolates (Table 1; also see Fig. S1 in the supplemental material) . No significant difference was observed between the two methods in predicting three (ABC combination) or four (ABCC combination) EPIYA repeats. However, our method was 100% accurate in all cases of identification of cagA-negative strains, as well as predicting four or five EPIYA repeats (Table 1). In addition, when we compared the two techniques in five cases of H. pylori isolates containing two subclones with different number of EPIYA repeats, isolated from the same patient, our method accurately identified the presence of these subclones and furthermore predicted the right number of EPIYA repeats in all cases (Table 2). These results suggest that our prediction method based upon the size of PCR amplicons is quite robust since it can give accurate predictions, especially in the presence of multiple infective subclones expressing CagA protein with different numbers of EPIYA motifs.
TABLE 1.
Comparison between PCR methods for the correct determination of the number of EPIYA motifs within CagA in single clone isolates
| No. of EPIYA motifsa | No. of strains tested | No. of correct predictions
|
|
|---|---|---|---|
| This study | Argent et al. (7) | ||
| 0 | 6 | 6 | 2 |
| 2 | 1 | 1 | 0 |
| 3 | 9 | 9 | 9 |
| 4 | 13 | 13 | 12 |
| 5 | 3 | 3 | 2 |
As determined by sequencing.
TABLE 2.
Determination of EPIYA motifs within CagA in mixed isolates containing multiple closely related subclones isolated from the same host
| Case | Accession no.a | EPIYA status
|
|
|---|---|---|---|
| This study | Argent et al. (7) | ||
| alcag7 | AM295790-1 | ABC-ABCC | ABCC |
| alcag40 | AM295577-8 | ABC-ABCC | ABCC |
| alcag4 | AM292554-5 | ABC-ABCCC | ABCCC |
| Hp77 | AM292593-4 | AB-ABC | No pattern |
| eva9 | AM292596-7 | AB-ABC | ABC |
That is, the GenBank/EMBL/DDBJ accession numbers of individual paired subclones.
Differences in the number of EPIYA motifs within CagA protein in H. pylori isolates from children and adults.
We classified the population of cagA-positive strains with regards to EPIYA number and type (Table 3). The overwhelming majority of our clinical strains were found to harbor the ABC combination of EPIYA motifs (adults [67.3%], children [72.1%]; Table 3). In 10 strains isolated from adults (17.2%) and 7 strains from children (16.3%) we determined the ABCC combination of EPIYA motifs, as a result of a 34-amino-acid repetition including the EPIYA-C (Fig. 4). The absence of the EPIYA-B motif was detected in only one isolate derived from children (Hp51, AM292592). Finally, exclusively within adults, two strains were found to harbor five EPIYA motifs in the ABCCC combination and one strain the ABABC combination (Table 3 and Fig. 4). No other combinations of TPM-A, -B, or -C were observed within our representative sample population. With regard to the multiple infections, six cases (three adult and three children) involved subclones expressing CagA protein with three and four EPIYA motifs in the ABC-ABCC combination and two cases with AB-ABC combination (Table 3). Interestingly, as observed in the case of single-clone isolates, subclones derived from multiple infections, expressing CagA protein with five or more EPIYA motifs, were detected only within adults. These data suggest that although no qualitative or quantitative difference exists between the strains isolated from adults and children, with regard to three or four EPIYA motifs, strains expressing CagA protein with five or more EPIYA motifs were detected exclusively within the adult population.
TABLE 3.
CagA diversity with regard to the number and type of EPIYA motifs in cagA-positive clinical isolates
| Strain group and no. of EPIYA motifs | Type(s) of EPIYA motif | Adults (n = 58)a
|
Children (n = 43)a
|
||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Single strains | |||||
| 2 | AC | 0 | 0 | 1 | 2.3 |
| 3 | ABC | 39 | 67.3 | 31 | 72.1 |
| 4 | ABCC | 10 | 17.2 | 7 | 16.3 |
| 5 | ABCCC | 2 | 3.5 | 0 | 0 |
| 5 | ABABC | 1 | 1.7 | 0 | 0 |
| Mixed isolates | |||||
| 2 and 3 | AB-ABC | 1 | 1.7 | 1 | 2.3 |
| 3 and 4 | ABC-ABCC | 3 | 5.2 | 3 | 7.0 |
| 3 and 5 | ABC-ABCCC | 1 | 1.7 | 0 | 0 |
| 4 and 6 | ABCC-ABCCCC | 1 | 1.7 | 0 | 0 |
Percentages refer to the total number of cagA-positive isolates within each population. n, number of isolates.
DISCUSSION
In the present study, we proposed a simple strategy by which accurate prediction of the cagA status, as well as the number and type of EPIYA motifs, involves just one single-step PCR amplification of the region encoding for the EPIYA motifs in the cagA gene. After H. pylori isolation from the gastric biopsy and DNA extraction, samples were subjected to EPIYA PCR. We showed that the absence of EPIYA PCR amplicon can accurately identify cagA-negative cases and therefore no prior characterization of the cagA status of the isolates is needed. In the event of a PCR amplicon with sizes of 470 or 570 bp we could safely assume the presence of an ABC or an ABCC combination of EPIYA motifs within CagA. We verified the accuracy of such predictions concerning three or four EPIYA motifs by sequencing in 93 cases of the 101 cagA-positive isolates analyzed within our study. More specifically, 70 of 101 cagA-positive strains carried three EPIYA motifs arranged in the ABC combination, 17 strains harbored four EPIYA motifs in the ABCC combination, and six cases involved mixed infections of the ABC-ABCC type. Therefore, in the overwhelming majority of cases (92%), the size of the PCR amplicon can safely predict the number and type of EPIYA motifs present. However, in the case of EPIYA PCR amplicons at 370 or 670 bp (8% of the cases) corresponding to CagA species with two EPIYA repeats (AB or AC combination) or more than four EPIYA repeats (ABCCC or ABABC combination), sequencing was required to establish the exact type of EPIYA motif combination, although prediction of the correct number of EPIYA motifs was possible. Moreover, our approach enabled us to accurately predict the number of EPIYA motifs in cases of mixed infections, where closely related subclones bearing the same RAPD profile, but divergent numbers of EPIYA motifs, were isolated from the same host. It is a common observation that such a pool of H. pylori clones may exist in a dynamic equilibrium within potentially all H. pylori-positive hosts (11) and reflect the continuous selective environmental pressure created by acidity and individual host immune responses, as well as factors relating to antibiotic consumption and diet (10, 31). Such H. pylori isolates may contain two or more closely related subclones bearing the same RAPD profile and therefore be indistinguishable in common clinical practice. This is particularly important, since these divergent CagA species were shown to be normally expressed (5; the present study) and can induce various degrees of hummingbird phenotype upon infection of gastric epithelial cells (5). Therefore, by applying our strategy, we could detect the presence of such subclones with divergent EPIYA motifs and isolate them. However, it does not discriminate between coinfecting strains with different genotypes harboring CagA with the same number of EPIYA motifs.
Our approach is especially suited to clinical microbiology laboratories where H. pylori isolation from gastric biopsies is often made by the sweeping method and therefore the presence of multiple closely related subclones within an H. pylori isolate is possible. On the contrary, the presence of such subclones may severely weaken the specificity of the previously proposed PCR approach for the determination of the EPIYA motifs using primers specific to the EPIYA-A, -B, or -C coding sequences (7). Indeed, in our hands, such a technique was accurate only in the case of single-clone isolates, and it proved less powerful in cases of mixed infections. Finally, our proposed scheme involves only a single-step PCR and therefore is more cost-effective than the already published method which utilizes three different primer sets.
Based upon the positivity rates of our PCR amplification assay, we determined cagA presence in 75% of the adult and children populations analyzed. Our data are in line with CagA seroprevalence in Greece, which has been reported at 77.4% and was found to be constant across gender and age (4). More than 92% of the cagA-positive strains harbored a CagA protein with three or four EPIYA motifs arranged in the ABC or ABCC combination, respectively. We found no Eastern type strains circulating within our population, although our primers were designed to also detect such cases. The presence of EPIYA-A and -B motifs was detected in all our cagA-positive isolates, with the exception of only one strain harboring an AC EPIYA motif. Furthermore, we observed no differences between adults and children with reference to the distribution of strains expressing CagA with one or two EPIYA-C repeats. It was, however, exclusively within the adult population that we detected the presence of strains with more than five EPIYA motifs (5 isolates out of 135 strains analyzed). A much higher number of cases should be analyzed overall in order to perform robust statistical analysis and compare the prevalence of such strains in adults and children. However, our findings may suggest that cagA-positive strains can acquire additional EPIYA-C motifs throughout adulthood and thus contribute to H. pylori pathogenesis at a later age. H. pylori have been shown to exhibit unique genetic variability through out chronic H. pylori infection, and these genetic changes may be directed by homologous recombination (19).
We have developed a rapid approach for the accurate characterization of EPIYA motifs within the carboxyl terminus of CagA protein, even in the presence of multiple infecting subclones and successfully typed H. pylori strains from Greek adults and children. We are applying our approach to the study of the potential association of CagA divergence within the carboxyl-terminal end and the clinical outcome of the H. pylori-associated disease. Further analysis of our strains with respect to their ability to successfully translocate the CagA protein and interact with intracellular effectors within the gastric epithelial cells, through phosphorylation-dependent and -independent interactions, may provide more clues regarding their true pathogenic potential.
Supplementary Material
Acknowledgments
We thank the following physicians for collecting the gastric biopsies: S. Michopoulos, Gastroenterology Clinic, Alexandras General Hospital, Athens, Greece; G. J. Mantzaris, First Department of Gastroenterology, Evangelismos Hospital, Athens, Greece; E. Katsiyiannaki, Pediatric Gastroenterology Unit, Aghia Sophia Children's Hospital, Athens, Greece; and E. Roma, First Department of Pediatrics, Athens University School of Medicine, Athens, Greece.
This project was funded by an internal grant from Hellenic Pasteur Institute and EGP is a postgraduate student supported by a Ph.D. studentship from the Hellenic Pasteur Institute.
Footnotes
Published ahead of print on 6 December 2006.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol. 28:37-53. [DOI] [PubMed] [Google Scholar]
- 2.Akopyanz, N., N. O. Bukanov, T. U. Westblom, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amieva, M. R., R. Vogelmann, A. Covacci, L. S. Tompkins, W. J. Nelson, and S. Falkow. 2003. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science 300:1430-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apostolopoulos, P., I. Vafiadis-Zouboulis, M. Tzivras, D. Kourtessas, N. Katsilambros, and A. Archimandritis. 2002. Helicobacter pylori (H pylori) infection in Greece: the changing prevalence during a ten-year period and its antigenic profile. BMC Gastroenterol. 2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aras, R. A., Y. Lee, S. K. Kim, D. Israel, R. M. Peek, Jr., and M. J. Blaser. 2003. Natural variation in populations of persistently colonizing bacteria affect human host cell phenotype. J. Infect. Dis. 188:486-496. [DOI] [PubMed] [Google Scholar]
- 6.Argent R. H., M. Kidd, R. J. Owen, R. J. Thomas, M. C. Limb, and J. C. Atherton. 2004.. Determinants and consequences of different levels of CagA phosphorylation for clinical isolates of Helicobacter pylori. Gastroenterology 127:514-523. [DOI] [PubMed] [Google Scholar]
- 7.Argent, R. H., Y. Zhang, and J. C. Atherton. 2005. Simple method for determination of the number of Helicobacter pylori CagA variable-region EPIYA tyrosine phosphorylation motifs by PCR. J. Clin. Microbiol. 43:791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azuma, T., S. Yamazaki, A. Yamakawa, M. Ohtani, A. Muramatsu, H. Suto, Y. Ito, M. Dojo, Y. Yamazaki, M. Kuriyama, Y. Keida, H. Higashi, and M. Hatakeyama. 2004. Association between diversity in the Src homology 2 domain-containing tyrosine phosphatase binding site of Helicobacter pylori CagA protein and gastric atrophy and cancer. J. Infect. Dis. 189:820-827. [DOI] [PubMed] [Google Scholar]
- 9.Backert, S., S. Moese, M. Selbach, V. Brinkmann, and T. F. Meyer. 2001. Phosphorylation of tyrosine 972 of the Helicobacter pylori CagA protein is essential for induction of a scattering phenotype in gastric epithelial cells. Mol. Microbiol. 42:631-644. [DOI] [PubMed] [Google Scholar]
- 10.Bjorkholm, B., A. Lundin, A. Sillen, K. Guillemin, N. Salama, C. Rubio, J. I. Gordon, P. Falk, and L. Engstrand. 2001. Comparison of genetic divergence and fitness between two subclones of Helicobacter pylori. Infect. Immun. 69:7832-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaser, M. J., and J. C. Atherton. 2004. Helicobacter pylori persistence: biology and disease. J. Clin. Investig. 113:321-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blaser, M. J., G. I. Perez-Perez, H. Kleanthous, T. L. Cover, R. M. Peek, P. H. Chyou, G. N. Stemmermann, and A. Nomura. 1995. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 55:2111-2115. [PubMed] [Google Scholar]
- 13.Churin, Y., L. Al-Ghoul, O. Kepp, T. F. Meyer, W. Birchmeier, and M. Naumann. 2003. Helicobacter pylori CagA protein targets the c-Met receptor and enhances the motogenic response. J. Cell Biol. 161:249-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Covacci, A., S. Censini, M. Bugnoli, R. Petracca, D. Burroni, G. Macchia, A. Massone, E. Papini, Z. Xiang, N. Figura, and R. Rappuoli. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. USA 90:5791-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crabtree, J. E., J. I. Wyatt, G. M. Sobala, G. Miller, D. S. Tompkins, J. N. Primrose, and A. G. Morgan. 1993. Systemic and mucosal humoral responses to Helicobacter pylori in gastric cancer. Gut 34:1339-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higashi, H., R. Tsutsumi, S. Muto, T. Sugiyama, T. Azuma, M. Asaka, and M. Hatakeyama. 2002. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science 295:683-686. [DOI] [PubMed] [Google Scholar]
- 17.Higashi, H., R. Tsutsumi, A. Fujita, S. Yamazaki, M. Asaka, T. Azuma, and M. Hatakeyama. 2002. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc. Natl. Acad. Sci. USA 99:14428-14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higashi, H., K. Yokoyama, Y. Fujii, S. Ren, H. Yuasa, I. Saadat, N. Murata-Kamiya, T. Azuma, and M. Hatakeyama. 2005. EPIYA motif is a membrane-targeting signal of Helicobacter pylori virulence factor CagA in mammalian cells. J. Biol. Chem. 280:23130-23137. [DOI] [PubMed] [Google Scholar]
- 19.Kraft, C., A. Stack, C. Josenhans, E. Niehus, G. Dietrich, P. Correa, J. G. Fox, D. Falush, and S. Suerbaum. 2006. Genomic changes during chronic Helicobacter pylori infection. J. Bacteriol. 188:249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuipers, E. J., G. I. Perez-Perez, S. G. Meuwissen, and M. J. Blaser. 1995. Helicobacter pylori and atrophic gastritis: importance of the cagA status. J. Natl. Cancer Inst. 87:1777-1780. [DOI] [PubMed] [Google Scholar]
- 21.Mimuro, H., T. Suzuki, J. Tanaka, M. Asahi, R. Haas, and C. Sasakawa. 2002. Grb2 is a key mediator of Helicobacter pylori CagA protein activities. Mol. Cell 10:745-755. [DOI] [PubMed] [Google Scholar]
- 22.Naito, M., T. Yamazaki, R. Tsutsumi, H. Higashi, K. Onoe, S. Yamazaki, T. Azuma, and M. Hatakeyama. 2006. Influence of EPIYA-repeat polymorphism on the phosphorylation-dependent biological activity of Helicobacter pylori CagA. Gastroenterology 130:1181-1190. [DOI] [PubMed] [Google Scholar]
- 23.Nomura, A. M., G. I. Perez-Perez, J. Lee, G. Stemmermann, and M. J. Blaser. 2002. Relationship between H. pylori cagA status and risk of peptic ulcer disease. Am. J. Epidemiol. 155:1054-1059. [DOI] [PubMed] [Google Scholar]
- 24.Odenbreit, S., J. Puls, B. Sedlmaier, E. Gerland, W. Fischer, and R. Haas. 2000. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287:1497-1500. [DOI] [PubMed] [Google Scholar]
- 25.Parsonnet, J., G. D. Friedman, N. Orentreich, and H. Vogelman. 1997. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut 40:297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudi, J., C. Kolb, M. Maiwald, D. Kuck, A. Sieg, P. R. Galle, and W. Stremmel. 1998. Diversity of Helicobacter pylori vacA and cagA genes and relationship to VacA and CagA protein expression, cytotoxin production, and associated diseases. J. Clin. Microbiol. 36:944-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segal, E. D., J. Cha, J. Lo, S. Falkow, and L. S. Tompkins. 1999. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. USA 96:14559-14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selbach, M., S. Moese, C. R. Hauck, T. F. Meyer, and S. Backert. 2002. Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo. J. Biol. Chem. 277:6775-6778. [DOI] [PubMed] [Google Scholar]
- 29.Selbach, M., S. Moese, R. Hurwitz, C. R. Hauck, T. F. Meyer, and S. Backert. 2003. The Helicobacter pylori CagA protein induces cortactin dephosphorylation and actin rearrangement by c-Src inactivation. EMBO J. 22:515-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stein, M., F. Bagnoli, R. Halenbeck, R. Rappuoli, W. J. Fantl, and A. Covacci. 2002. c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol. Microbiol. 43:971-980. [DOI] [PubMed] [Google Scholar]
- 31.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 33.van Doorn, L. J., C. Figueiredo, R. Sanna, M. J. Blaser, and W. G. Quint. 1999. Distinct variants of Helicobacter pylori cagA are associated with vacA subtypes. J. Clin. Microbiol. 37:2306-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reference deleted.
- 35.Yamaoka, Y., T. Kodama, K. Kashima, D. Y. Graham, and A. R. Sepulveda. 1998. Variants of the 3′ region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J. Clin. Microbiol. 36:2258-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou, W., S. Yamazaki, A. Yamakawa, M. Ohtani, Y. Ito, Y. Keida, H. Higashi, M. Hatakeyama, J. Si, and T. Azuma. 2004. The diversity of vacA and cagA genes of Helicobacter pylori in East Asia. FEMS Immunol. Med. Microbiol. 40:81-87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.