Abstract
We evaluated three multilocus sequence typing (MLST) schemes for Staphylococcus epidermidis and selected the seven most discriminatory loci for the formation of a new, more powerful MLST scheme. This improved scheme gave 31 sequence types (STs) and 5 clonal complexes (CCs), whereas the other schemes delineate 16 to 24 STs and 1 to 3 CCs.
Two different multilocus sequence typing (MLST) schemes were published for Staphylococcus epidermidis in 2003 (9, 10). Wisplinghoff et al. (10) (data previously held on www.mlst.net) and Wang et al. (9) described S. epidermidis MLST schemes that were largely based on the scheme for Staphylococcus aureus (2). By the use of different isolate collections, neither scheme was able to discriminate more than two clonal complexes (CCs). A third unpublished scheme, developed by S. J. Peacock et al., used two S. aureus MLST loci, gmk and pta, together with five new loci, dfp, gtr, mutS, pyrR, and xpt. Three separate MLST protocols for the typing of S. epidermidis are clearly unsatisfactory, especially as none of these methods provides a high level of discrimination. Here we assess the ability of the 14 different loci used in these three schemes to discriminate between isolates of S. epidermidis.
Bacterial strains and media.
A collection of 66 geographically diverse S. epidermidis isolates was analyzed in this study (10). Of these, 46 isolates were recovered from the blood of patients with prosthetic valve endocarditis (PVE), and 20 isolates were from bloodstream infections (BSIs) that were not linked to PVE. Isolates were grown overnight at 37°C on tryptic soy agar (Oxoid, Basingstoke, United Kingdom) and were stored at −80°C in 15% (vol/vol) glycerol and tryptic soy broth (Oxoid).
Multilocus sequence typing.
All isolates were analyzed by three MLST protocols, as previously described by Wisplinghoff et al. (10), Wang et al. (9), and Peacock et al. (the primer sequences are provided in Table 1). Allele 1 of yqiL described in the scheme devised by Wang et al. (9) was lengthened to coincide with the fragment size of the remaining alleles at that locus. Additional sequences were acquired as necessary from the published genome of S. epidermidis RP62A (3). Chromosomal DNA was isolated with DNeasy kits (QIAGEN). PCR was performed with 29-μl reaction volumes, comprising 1 μl of each primer (10 pmol) and 25 μl of ReddyMix master mix (ABgene, Epsom, United Kingdom). The amplicons were further purified and sequenced. The sequences of both strands of all PCR products were resolved with an ABI 3700 automated sequencer (PE Applied Biosystems) with BigDye (version 3) fluorescent terminators and the primers used in the initial PCR amplification.
TABLE 1.
Modified primer sequences for an improved S. epidermidis MLST scheme
| Gene | Primer | Sequence (5′-3′) |
|---|---|---|
| arcC | arcC-F | TGTGATGAGCACGCTACCGTTAG |
| arcC-R | TCCAAGTAAACCCATCGGTCTG | |
| aroE | aroE-F | CATTGGATTACCTCTTTGTTCAGC |
| aroE-R | CAAGCGAAATCTGTTGGGG | |
| gtr | gtr-F | CAGCCAATTCTTTTATGACTTTT |
| gtr-R | GTGATTAAAGGTATTGATTTGAAT | |
| mutS | mutS-F3 | GATATAAGAATAAGGGTTGTGAA |
| mutS-R3 | GTAATCGTCTCAGTTATCATGTT | |
| pyrR | pyr-F2 | GTTACTAATACTTTTGCTGTGTTT |
| pyr-R4 | GTAGAATGTAAAGAGACTAAAATGAA | |
| tpi | tpi-F2 | ATCCAATTAGACGCTTTAGTAAC |
| tpi-R2 | TTAATGATGCGCCACCTACA | |
| yqiL | yqiL-F2 | CACGCATAGTATTAGCTGAAG |
| yqiL-R2 | CTAATGCCTTCATCTTGAGAAATAA | |
| dfpFa | dfpF-F | TAAGAGTTATGCTATCTGATCAT |
| dfpF-R | GGATCAATAACTTCAACTGTAG | |
| gmka | gmk-F | GACTGTTAATTGTTCTTTCAGG |
| gmk-R | GTTAACTACAACGTAGTCGTAT | |
| ptaa | pta-F | GTAAAAATAGTATTACCTGAAGG |
| pta-R | GAACCTTTTGTAGAAAAGCTTAA | |
| xpta | xpt-F | ATTTGCTAAAAAAGCTAAACTA |
| xpt-R | CTTTATTTGCTTGAAGAAATGTC |
The primer sequences used in the method of Peacock et al. but not present in the improved scheme.
The alleles at each locus were distinguished by using Sequence Output software (available from http://www.mlst.net). Allelic profiles consisting of the allele numbers at each of the seven loci were assigned to sequence types (STs) for each of three MLST schemes, as described previously (7).
Use of eBURST algorithm.
Lineages were analyzed separately for each MLST scheme by using the eBURST algorithm (5), available at http://eBURST.mlst.net. Clonal complexes were defined by using the default setting, in which all STs within a clonal complex differ by no more than one allele from at least one other ST in the clonal complex, as recommended by Spratt and colleagues (5).
Simpson's D.
Simpson's index of diversity (D) values for each MLST scheme were calculated as described by Hunter and Gaston (6). The 95% confidence intervals were calculated as described by Grundmann et al. (4).
MLST scheme comparison.
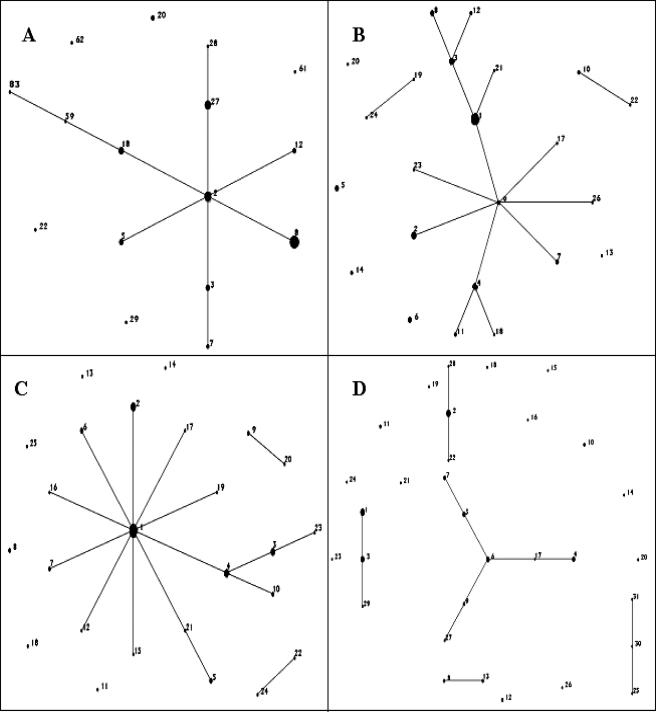
Comparison of Simpson's index of diversity values for all three MLST schemes revealed that the format proposed by Peacock et al. proved to be the most discriminatory, with D equal to 0.922 (compared to D values of 0.889 and 0.871 for the schemes of Wang et al. [9] and Wisplinghoff et al. [10], respectively), although this was not significant (P < 0.05) (Table 2). The three schemes resolved between 15 STs (scheme of Wisplinghof et al. [10]) and 25 STs (scheme of Wang et al. [9]); the scheme of Peacock et al. resolved 23 STs. The MLST schemes of both Peacock et al. and Wang et al. (9) split the isolates into three separate clonal complexes, while the scheme of Wisplinghoff et al. (10) generated a single large clonal complex (Fig. 1).
TABLE 2.
Summary of statistics for all loci and MLST schemes
| MLST scheme and locusa | Length of sequenced fragment (bp) | No. of alleles | No. of polymorphic sites | D | 95% CIb |
|---|---|---|---|---|---|
| Peacock et al. | 0.922 | 0.884-0.960 | |||
| dfp | 456 | 7 | 12 | 0.398 | 0.250-0.548 |
| gmk | 405 | 5 | 12 | 0.345 | 0.203-0.487 |
| gtr | 438 | 6 | 15 | 0.585 | 0.460-0.710 |
| mutS | 412 | 6 | 7 | 0.552 | 0.432-0.671 |
| pta | 410 | 5 | 11 | 0.398 | 0.267-0.529 |
| pyrR | 428 | 8 | 16 | 0.744 | 0.679-0.808 |
| xpt | 520 | 6 | 13 | 0.347 | 0.203-0.491 |
| Wang et al. (9) | 0.889 | 0.849-0.950 | |||
| aroE | 482 | 7 | 32 | 0.327 | 0.180-0.473 |
| glpF | 450 | 7 | 15 | 0.394 | 0.248-0.541 |
| gmk | 391 | 5 | 12 | 0.322 | 0.181-0.463 |
| hsp60 | 421 | 6 | 12 | 0.228 | 0.091-0.364 |
| pta | 463 | 7 | 13 | 0.430 | 0.290-0.569 |
| tpi | 424 | 7 | 11 | 0.534 | 0.424-0.644 |
| yqiL | 416 | 6 | 12 | 0.558 | 0.446-0.669 |
| Wisplinghoff et al. (10) | 0.871 | 0.821-0.920 | |||
| arc | 465 | 5 | 15 | 0.574 | 0.456-0.693 |
| aroE | 420 | 6 | 16 | 0.570 | 0.474-0.667 |
| glpK | 468 | 6 | 16 | 0.201 | 0.070-0.332 |
| gmk | 465 | 5 | 12 | 0.345 | 0.203-0.487 |
| pta | 477 | 4 | 11 | 0.356 | 0.224-0.489 |
| tpi | 408 | 6 | 8 | 0.227 | 0.091-0.362 |
| yqiL | 474 | 4 | 14 | 0.172 | 0.050-0.294 |
| Improved MLST scheme | 0.956 | 0.935-0.976 |
Loci highlighted in boldface indicate those used in the unified MLST scheme.
CI, confidence interval.
FIG. 1.
eBURST representation of MLST data for (A) the MLST scheme of Wisplinghoff et al. (10), (B) the MLST scheme of Peacock et al., (C) the MLST scheme of Wang et al. (9), and (D) the improved MLST scheme.
A single improved MLST scheme.
An MLST scheme that consisted of the seven most discriminatory alleles from the three schemes was developed (Table 2). PCR involved an initial denaturation of 95°C for 3 min; 34 cycles of 95°C for 30 s, 50°C for 1 min, and 72°C for 1 min; and a final extension of 72°C for 10 min. The modified MLST scheme was more discriminatory than the scheme of Wisplinghoff et al. (10) (P < 0.05), with D equal to 0.956, but was not significantly more discriminatory than either the MLST scheme of Peacock et al. or that of Wang et al. (9). However, it did resolve the isolate collection into 31 STs and 5 distinct clonal complexes (Fig. 1) and more singletons than the other schemes.
Clinical disease.
All STs with the exception of ST2 and ST7 contained isolates from patients with either BSIs or PVE; ST2 and ST7 contained examples of isolates from both types of patients. STs 5, 11, 15, 16, 18, 19, 20, 21, 22, 23, and 24 contained only isolates from patients with BSIs; and the remaining 18 STs contained only isolates from patients with PVE.
The STs resolved by the improved MLST scheme correlated in all but two cases with the isolates that caused either a bloodstream infection or prosthetic valve endocarditis, while three of the five clonal complexes were associated with only prosthetic valve endocarditis. In comparison, two of the three CCs generated by the scheme of Peacock et al. (CC9 and CC10) included strains associated with both PVE and BSIs, while the main clonal complex generated by the scheme of Wang et al. (9) also possessed strains isolated from both types of patients. One of the clonal complexes from the new MLST scheme (CC2) that included isolates from both types of patients also included one of the two STs associated with both BSIs and PVE as its ancestral strain (ST2), which has two single-locus variants (SLVs). Of these two SLVs, one, ST28, was from a patient with PVE, while the other, ST22, was from a patient with a bloodstream infection. It is our hypothesis that this clonal complex represents the branching of two distinct lineages, one linked with PVE and one linked with BSIs.
While none of the MLST schemes presented here is as discriminatory as those developed for other species, including S. aureus (D = 0.996) (2), Streptococcus pneumoniae (D = 0.998) (1), Neisseria meningitidis (D = 0.995) (7), and Haemophilus influenzae (D = 0.996) (8), we believe that the improved MLST scheme detailed here provides the best discrimination of the MLST formats currently available for the typing of S. epidermidis strains.
Nucleotide sequence accession numbers.
The novel gene sequences reported in this study have been deposited in GenBank (accession numbers DQ991011 to DQ991028).
Acknowledgments
This work was supported by the Wellcome Trust. M.C.E. is a Royal Society University Research Fellow. S.J.P. is a Wellcome Trust Career Develop Fellow in Clinical Tropical Medicine.
We thank Paul Wilkinson for technical assistance.
Footnotes
Published ahead of print on 6 December 2006.
REFERENCES
- 1.Enright, M., and B. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 2.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. DeBoy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 101:9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meats, E., E. J. Feil, S. Stringer, A. J. Cody, R. Goldstein, J. S. Kroll, T. Popovic, and B. G. Spratt. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 41:1623-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, X. M., L. Noble, B. N. Kreiswirth, W. Eisner, W. McClements, K. U. Jansen, and A. S. Anderson. 2003. Evaluation of a multilocus sequence typing system for Staphylococcus epidermidis. J. Med. Microbiol. 52:989-998. [DOI] [PubMed] [Google Scholar]
- 10.Wisplinghoff, H., A. E. Rosato, M. C. Enright, M. Noto, W. Craig, and G. L. Archer. 2003. Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob. Agents Chemother. 47:3574-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]