Abstract
The factors determining whether a person infected with Entamoeba histolytica develops disease remain obscure. To investigate whether the parasite genome contributes to the outcome, we have investigated the distribution of parasite genotypes among E. histolytica-infected individuals in Bangladesh. Samples were obtained from individuals who either were asymptomatic, had diarrhea/dysentery, or had developed a liver abscess. Genotypes were determined by using six tRNA-linked polymorphic markers, and their distributions among the three sample groups were evaluated. A significant population differentiation in the genotype distribution was found for four of the six individual markers as well as for the combined genotypes, suggesting that the parasite genome does contribute in some way to the outcome of infection with E. histolytica. The markers themselves do not indicate the nature of the underlying genetic differences, but they may be linked to loci that do have an impact on the outcome of infection.
Amebiasis is one of the leading causes of death due to parasitic infection worldwide (15), but it has long been known that not all Entamoeba histolytica infections lead to disease. This was first observed almost a century ago in subjects experimentally infected in the Philippines (17) and has been confirmed repeatedly in more recent times (3, 10, 16, 18). Indeed, less than 1 in 10 infections are now thought to result in intestinal or extraintestinal symptoms in humans. The variables that are responsible for determining the different outcomes of infection are still largely unknown. At present we do not know whether some E. histolytica strains are intrinsically more virulent than others, but it is possible that the genetic makeup of the parasite is one contributing factor.
In order to investigate whether there is any link between the parasite and the outcome of infection, a reliable method for genotyping the organism is required. A number of methods for detecting diversity in E. histolytica have been described over the years (4), but we recently described a PCR-based approach that is highly sensitive and discriminatory (1). The six targets for amplification in this method were selected from among the 40-plus short tandem repeat (STR)-containing loci linked to tRNA genes in E. histolytica (5). In the present study we applied this genotyping system to a panel of samples isolated from Bangladeshi individuals who either were asymptomatic or had intestinal or extraintestinal disease. Statistical analyses of the results suggest that the parasite genome plays a role in determining the outcome of infection with E. histolytica.
MATERIALS AND METHODS
E. histolytica samples.
Three groups of samples from Bangladeshi individuals with E. histolytica infections were collected. All the “asymptomatic” and “diarrhea/dysentery” samples were from an ongoing field project studying preschool children (2 to 5 years old) for immunity to amebiasis in Mirpur, about 5 miles from Dhaka, Bangladesh (12). As part of this study, stool specimens are obtained every month for detection of E. histolytica infection by a stool antigen-capture enzyme-linked immunosorbent assay and culture. If diarrheal disease develops, a stool sample is obtained and studied for enteropathogens and the child is examined. All samples are transported to the International Centre for Diarrheal Disease Research, Bangladesh (ICDDR,B), laboratory for processing within 2 to 4 h.
Entamoeba histolytica infection is defined by a positive test result for amebic antigen in stool. A “new episode” of E. histolytica infection during the period of observation is defined as a positive E. histolytica stool antigen and/or culture result preceded by two or more monthly surveillance stool samples with negative results. This definition is based on that used in earlier studies with this cohort that demonstrated a requirement for two negative monthly stool samples to obtain the highest separation between new and relapsed infections, as judged by DNA fingerprinting of the parasite by serine-rich E. histolytica protein (SREHP) gene polymorphism (11).
E. histolytica-associated diarrhea is defined as three or more unformed stools in a 24-h period accompanying a new episode of E. histolytica infection. This definition has previously been validated with this cohort by demonstrating that diarrhea was approximately five times more common in the setting of a new infection (age-adjusted odds ratio for the association of new E. histolytica infection with diarrhea, 4.7; 95% confidence interval, 2.9 to 7.6) (13). Amebic dysentery is defined as a diarrheal stool sample containing occult or gross blood that was positive for E. histolytica antigen (13). Coinfection with Shigella or Campylobacter in children with E. histolytica-associated diarrhea was observed in only 9% (6/69) of the episodes for which a complete microbiologic workup was performed (12). In contrast, a complete workup for enteropathogens in 210 episodes of non-E. histolytica-associated diarrhea identified an etiologic agent in 59% of cases (13). This indicates that the symptoms in patients with E. histolytica-associated diarrhea, as defined above, are strongly linked to the presence of the ameba.
Amebic liver abscess (ALA) samples were from patients admitted to various clinics and hospitals in Dhaka. Liver abscesses are routinely aspirated and the aspirated pus is sent to ICDDR,B for diagnostic purposes. The use of human subjects was approved by the Ethical Review Committee of the ICDDR,B and the Human Investigation Committee of the University of Virginia. Informed consent was obtained from the parents of the children.
The organisms in a total of 155 E. histolytica-positive samples, as determined by amebic stool antigen detection (E. histolytica II kit; TechLab, Inc., Blacksburg, VA) and/or small-subunit rRNA gene amplification (14), were typed in the present study.
In some cases multiple samples were available from the same child. If the organisms in the follow-up samples (i.e., the subsequent monthly sample or a diarrheal sample) showed no change in genotype, only one sample was included in the statistical analysis. If organisms of the same genotype were identified in both asymptomatic and diarrheal samples from the same child, the result was included in the diarrhea/dysentery group only.
For the diarrhea/dysentery group, only samples that met the stringent definitions (described above) of E. histolytica-associated diarrhea or amebic dysentery were included. Four samples that met the definitions but that had Campylobacter and/or Shigella coinfection were excluded, as the disease origin could not be definitively assigned to the ameba.
The final number of samples included in the statistical analysis was 111: 38 in the asymptomatic group, 30 in the diarrhea/dysentery group, and 43 in the liver abscess group.
Isolation of DNA and PCR amplification.
DNA from the culture, stool, or ALA pus lysates was purified by the cetyltrimethylammonium bromide extraction method or with a QIAquick DNA stool minikit (QIAGEN, Ltd., United Kingdom), as described previously (1). It has previously been shown that the establishment of amebae in culture has no effect on the genotype (19). PCR amplification was performed with E. histolytica-specific primer pairs targeting the tRNA-linked polymorphic STR loci, with BioTaq polymerase (Bioline Ltd., United Kingdom), and under the conditions already described (1). In a few cases a nested PCR was performed first with tRNA-specific primers and then with the E. histolytica-specific pair of primers to improve detection.
Genotype assignment.
PCR products were separated in 1.5% agarose gels as described previously (1). The resolving power of these gels is sufficient to detect differences in product sizes as small as 8 bp (verified by product sequencing [unpublished data]). Each unique amplification product size was assigned an “STR type” number at each locus (see Table S1 in the supplemental material). In cases in which the assignment was uncertain, the products were reanalyzed in lanes adjacent to reference samples. By using a combination of the assigned numbers for all six loci, each isolate was then assigned an overall genotype number.
Data analysis.
Statistical analysis of the genotype distribution among the three sample groups was performed by using the Arlequin software package (http://anthro.unige.ch/arlequin/ [8]). Both individual locus data and overall genotype data were analyzed. The total number of predicted genotypes was calculated by using the Chao estimator (7).
RESULTS
Genotypes based on PCR amplification patterns at six loci.
PCR amplification was successful for all samples, regardless of the origin of the DNA or the method of DNA isolation (Fig. 1). Although a single PCR product was the norm, in nine samples we observed a second faint band in the amplification of some loci; but for simplicity, we considered only the higher-intensity band for genotype assignment. Across all samples and by combining the results obtained with all six markers, a total of 26, 18, and 41 genotypes were identified in the asymptomatic, diarrhea/dysentery, and liver abscess sample groups, respectively. The number of distinct size variants at individual loci ranged from 3 to 10 (Table 1).
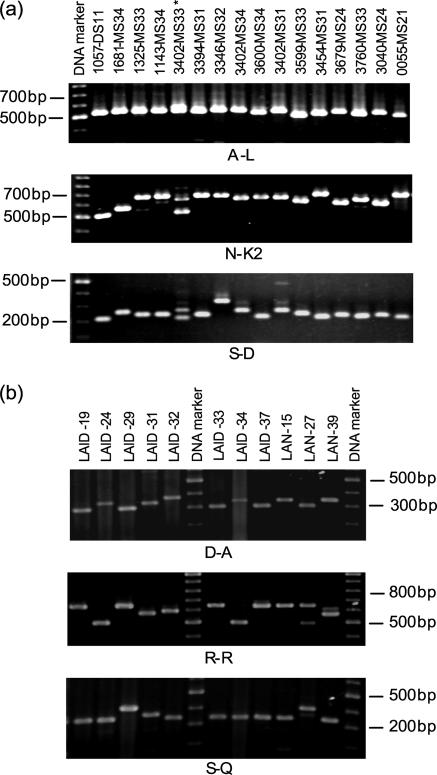
FIG. 1.
tRNA-linked STR patterns of a selection of E. histolytica samples. (a) Mirpur samples at the A-L, N-K2, and S-D loci. All samples except sample 1057-DS11 were from individuals with asymptomatic infections. *, transiently infected sample, as shown by double bands at the N-K2 and S-D loci. (b) Liver abscess samples at D-A, R-R, and S-Q loci.
TABLE 1.
Genotyping data and analyses
| Sample group | Locus | No. of samples showing STR typea:
|
Pairwise FST (sample group)b | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |||
| Asymptomatic | A-L | 5 | 29 | 3 | 1 | 0.030 (As-DD) | |||||||
| Diarrhea/dysentery | 27 | 3 | 0.088** (As-LA) | ||||||||||
| Liver abscess | 3 | 15 | 21 | 4 | 0.229** (DD-LA) | ||||||||
| Asymptomatic | D-A | 16 | 19 | 2 | 1 | 0.162** (As-DD) | |||||||
| Diarrhea/dysentery | 4 | 25 | 1 | −0.008 (As-LA) | |||||||||
| Liver abscess | 13 | 23 | 5 | 2 | 0.098** (DD-LA) | ||||||||
| Asymptomatic | N-K2 | 1 | 2 | 2 | 1 | 5 | 2 | 3 | 21 | 1 | 0.265** (As-DD) | ||
| Diarrhea/dysentery | 4 | 1 | 1 | 19 | 5 | 0.250** (As-LA) | |||||||
| Liver abscess | 4 | 1 | 3 | 8 | 6 | 1 | 20 | 0.049 (DD-LA) | |||||
| Asymptomatic | R-R | 4 | 1 | 11 | 17 | 5 | −0.026 (As-DD) | ||||||
| Diarrhea/dysentery | 4 | 8 | 15 | 3 | 0.003 (As-LA) | ||||||||
| Liver abscess | 11 | 8 | 16 | 7 | 1 | 0.001 (DD-LA) | |||||||
| Asymptomatic | S-D | 2 | 14 | 12 | 1 | 7 | 1 | 1 | 0.086 (As-DD) | ||||
| Diarrhea/dysentery | 9 | 20 | 1 | −0.007 (As-LA) | |||||||||
| Liver abscess | 1 | 3 | 13 | 14 | 5 | 3 | 1 | 1 | 1 | 1 | 0.070 (DD-LA) | ||
| Asymptomatic | S-Q | 5 | 21 | 2 | 5 | 2 | 3 | −0.020 (As-DD) | |||||
| Diarrhea/dysentery | 1 | 3 | 18 | 3 | 4 | 1 | 0.195** (As-LA) | ||||||
| Liver abscess | 1 | 5 | 6 | 21 | 2 | 3 | 1 | 1 | 2 | 1 | 0.207** (DD-LA) | ||
| Overall result | 0.094** (As-DD) | ||||||||||||
| 0.100** (As-LA) | |||||||||||||
| 0.107** (DD-LA) | |||||||||||||
Certain STR types were not detected in these Bangladeshi samples but have been found in samples from elsewhere in the world.
Sample groups: As, asymptomatic; DD, diarrhea/dysentery; and LA, liver abscess. FST, Wright's F statistic. FST values are the population pairwise FSTs (calculated by using the Arlequin software package), with significant population differentiation indicated by ** (P < 0.01).
In the asymptomatic group, the most common genotypes were 51 (four children) and 13 and 19 (three children each). In the diarrhea/dysentery group, the most common genotypes were 66 (seven patients) and 45 and 50 (three patients each). Only two genotypes, genotypes 23 and 39, were found in more than one ALA patient, and these were found in two patients each. The individual STR types making up these genotypes are given in Table S2A in the supplemental material. The households of the children infected with the most common genotype, genotype 66, had a widespread distribution in Mirpur.
Genotype distribution among sample groups.
In addition to the overall population comparison, the PCR patterns at two of the six loci (the D-A and N-K2 loci) showed a strong statistically significant difference in their distributions among the two sample groups from Mirpur (Table 1). In particular, two PCR patterns in locus D-A stand out: STR type 2, found in 16 of 38 samples from asymptomatic individuals but only 4 of 30 samples from patients with diarrhea/dysentery, and STR type 3, found in 25 of 30 samples from patients with diarrhea/dysentery but only 19 of 38 samples from asymptomatic individuals. Similarly, two PCR patterns in locus N-K2 appear to have contributed the most to this result: STR type 8, found in 19 of 30 samples from patients with diarrhea/dysentery but only 3 of 38 samples from asymptomatic individuals, and STR type 9, found in 21 of 38 samples from asymptomatic individuals but only 5 of 30 samples from patients with diarrhea/dysentery.
When the liver abscess samples were included, we found that the overall difference in genotypes between each of the two population pairs was highly significant (Table 1). Between the asymptomatic and the liver abscess sample groups, the loci showing the greatest difference in distribution were A-L (STR types 1 and 2), N-K2 (STR types 8 and 9), and S-Q (STR types 3 and 4), while between the diarrhea/dysentery and liver abscess groups, the greatest differences were again in loci A-L (STR types 2 and 3) and S-Q (STR types 3 and 4), with the addition of locus D-A (STR types 2 and 3). Variations at the R-R and S-D loci never showed significant differentiation between the sample groups.
Association between individual genotypes and outcome of infection.
The total number of genotypes was very high; therefore, the number of samples represented by any one genotype was low. We tested only the most common genotype for an association with the outcome of infection (see Table S2B in the supplemental material). Genotype 66 was significantly more common in samples from the diarrhea/dysentery group (P = 0.0005). Nevertheless, genotype 66 was found in only 7 of the 30 samples from the diarrhea/dysentery group (23.3%).
Predicted total number of genotypes.
The Chao estimator was used to predict the total number of genotypes in Bangladesh (7). Among the samples from the asymptomatic and diarrhea/dysentery groups, the total predicted number of genotypes in Mirpur children alone is 113, more than twice the number detected in this study. With the liver abscess samples added, the predicted number of genotypes across Bangladesh is 357. This is probably an underestimate, since sampling was not uniform or comprehensive.
E. histolytica follow-up samples.
Follow-up samples were available from 23 children, and we investigated the stabilities of the genotype patterns of the organisms from these infections. A maximum of seven follow-up samples were available from one child over a period of 11 months, and the same genotype was detected in all except one of the samples (sample 3402-MS33), which seemed to show a mixture of two genotypes (Fig. 1A). However the “new” genotype was not found in the subsequent monthly follow-up sample (sample 3402-MS34), suggesting that it was a transient infection. Transient infections have been reported previously (18). Six and five follow-up samples were available from one child each. Both of these children carried an infection with the same genotype throughout the follow-up period. Two and three follow-up samples were available from 13 and 7 children, respectively. The genotypes changed in 7 children, while in 13 children all the follow-up samples showed the same genotypes at all loci. On average, three of the six loci changed when a follow-up genotype was different, indicating that loss and reinfection are easily detectable. In four cases, an infection with the same genotype persisted for over 10 months; in each case the genotype was different, suggesting that the ability to produce a long-term infection is not restricted to a particular genotype.
DISCUSSION
Our main objective in this study was to evaluate the utility of our recently developed E. histolytica genotyping system in determining whether parasite genotypes correlate with the outcome of infection. We found evidence of a nonrandom distribution of parasite genotypes among the sample groups. However, there are some limitations in this study, which are described below, and these may have some impact on the observed population differentiation.
(i) All our asymptomatic and diarrhea/dysentery samples were from preschool children from Mirpur, and we do not know whether their genotypes are representative of those from all age groups and of the strain diversity across Bangladesh. All ALA samples were from adults in other areas of Dhaka. No adult or geographically matched asymptomatic or diarrhea/dysentery control groups were available, and so comparisons between the Mirpur and ALA samples must be interpreted with caution.
(ii) The genotypes were assigned on the basis of the estimated PCR product sizes in gels, and as a result some assignments could be incorrect. Nevertheless, sequencing of approximately half of the products at two loci (unpublished data) suggests that an incorrect STR type assignment is rare. The individual short tandem repeat units in the various loci are generally 8 or 9 bp in length, which means that a difference of one repeat more or less is usually clearly detectable under our gel conditions.
(iii) Our genotyping is based on PCR product size differences, but products of the same size do not necessarily have the same sequence. Sequencing has confirmed that this does occur. However, unless both sequence types show the same unequal distribution among sample populations, the combination of two different sequences in the same STR type is likely to reduce rather than increase any population differentiation signal that exists.
Using the six tRNA-linked STR loci, we detected 85 genotypes in 111 unrelated samples, a level of diversity which is comparable to that described in previous reports. Ayeh-Kumi et al. (2) also studied clinical samples from Bangladesh but used a nested PCR of the SREHP gene coupled with restriction digestion. They found 25 genotypes among 42 intestinal isolates and 9 genotypes among the isolates from 12 ALA samples. Eight of their nine genotypes among the liver abscess samples were unique to the ALA samples investigated. We also found that most of our ALA genotypes were unique and not detected among the intestinal strains. Clark and Diamond (6) observed 16 different genotypes among 18 isolates of E. histolytica from diverse geographical locations using the combined results of restriction digestion of SREHP and amplification of the SSG locus. Haghighi et al. (9) used sequencing of four loci (two tRNA-linked STR loci, chitinase, and SREHP) to investigate 79 isolates of E. histolytica, mostly from Japan and Thailand, but failed to find an association between the parasite genotype and the outcome of infection. The high level of diversity seen suggests that the rapid generation of new variants is occurring. If this is the case, then it may be that we have detected population differentiation only because our samples came from a geographically restricted population (Mirpur) and all of our samples were collected over a relatively short period of time (2 years).
The high degree of polymorphism detected in E. histolytica could have been problematic, but we did find a statistically significant difference in genotype representation among the three sample populations, suggesting that the parasite genome does influence the outcome of infection. Other approaches will be needed to identify the genes responsible for this result, as it is highly unlikely that the tRNA-linked STR loci are directly responsible in any way. It is also important to know whether these observations made in Bangladesh can be replicated in other parts of the world. To investigate this, extensive sample collections from additional areas of endemicity are needed.
Explaining the link between parasite genotype and infection outcome presents us with some theoretical problems. Existing evidence points to E. histolytica being a clonal organism; therefore, meiotic recombination and reassortment of genes should not be contributing to genotype differences. If this is true, a novel mechanism for generating genetic variation may exist. This could involve a recurring genome instability that leads to the repeated generation of the same genotypes through intragenomic reorganization and is reflected in turn in the tRNA-linked STR polymorphisms detected by our method. Only future genome-wide analyses of chromosome organization can address this possibility.
Supplementary Material
Acknowledgments
W. A. Petri, Jr., received research support from TechLab, Inc., and royalties from a patent license agreement with TechLab for a diagnostic test for amebiasis. These royalties accrue to the American Society of Tropical Medicine and Hygiene, without benefit to W.A. Petri, Jr. The other authors declare no conflicts of interest.
The genotyping was supported by grant 067314 from the Wellcome Trust, awarded to C. G. Clark. The Mirpur study is conducted at the ICDDR,B Centre for Health and Population Research with the support of a grant (grant AI-43596) from the National Institutes of Health through the University of Virginia. I. K. M. Ali was a doctoral student funded by the Commonwealth Scholarship Commission of Great Britain. R. Haque is a Howard Hughes Medical Institute International Research Scholar.
We thank the parents and children of Mirpur for their participation and Jian-Fen Shu for guidance on the statistical analysis.
Published ahead of print on 22 November 2006.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org.
REFERENCES
- 1.Ali, I. K. M., M. Zaki, and C. G. Clark. 2005. Use of PCR amplification of tRNA gene-linked short tandem repeats for genotyping Entamoeba histolytica. J. Clin. Microbiol. 43:5842-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayeh-Kumi, P. F., I. M. Ali, L. A. Lockhart, C. A. Gilchrist, W. A. Petri, Jr., and R. Haque. 2001. Entamoeba histolytica: genetic diversity of clinical isolates from Bangladesh as demonstrated by polymorphisms in the serine-rich gene. Exp. Parasitol. 99:80-88. [DOI] [PubMed] [Google Scholar]
- 3.Blessmann, J., I. K. M. Ali, P. A. Ton Nu, B. T. Dinh, T. Q. Ngo Viet, A. Le Van, C. G. Clark, and E. Tannich. 2003. Longitudinal study of intestinal Entamoeba histolytica infections in asymptomatic adult carriers. J. Clin. Microbiol. 41:4745-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark, C. G. 2006. Methods for the investigation of diversity in Entamoeba histolytica. Arch. Med. Res. 37:258-262. [DOI] [PubMed] [Google Scholar]
- 5.Clark, C. G., I. K. M. Ali, M. Zaki, B. J. Loftus, and N. Hall. 2006. Unique organisation of tRNA genes in Entamoeba histolytica. Mol. Biochem. Parasitol. 146:24-29. [DOI] [PubMed] [Google Scholar]
- 6.Clark, C. G., and L. S. Diamond. 1993. Entamoeba histolytica: a method for isolate identification. Exp. Parasitol. 77:450-455. [DOI] [PubMed] [Google Scholar]
- 7.Colwell, R. K., and J. A. Coddington. 1994. Estimating terrestrial biodiversity through extrapolation. Philos. Trans. R. Soc. Lond. B 345:101-118. [DOI] [PubMed] [Google Scholar]
- 8.Excoffier, L., G. Laval, and S. Schneider. 2005. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol. Bioinformat. Online 1:47-50. [PMC free article] [PubMed] [Google Scholar]
- 9.Haghighi, A., S. Kobayashi, T. Takeuchi, N. Thammapalerd, and T. Nozaki. 2003. Geographic diversity among genotypes of Entamoeba histolytica field isolates. J. Clin. Microbiol. 41:3748-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haque, R., I. M. Ali, and W. A. Petri, Jr. 1999. Prevalence and immune response to Entamoeba histolytica infection in preschool children in Bangladesh. Am. J. Trop. Med. Hyg. 60:1031-1034. [DOI] [PubMed] [Google Scholar]
- 11.Haque, R., P. Duggal, I. K. M. Ali, M. B. Hossain, D. Mondal, R. B. Sack, B. M. Farr, T. H. Beaty, and W. A. Petri, Jr. 2002. Innate and acquired resistance to amebiasis in Bangladeshi children. J. Infect. Dis. 186:547-552. [DOI] [PubMed] [Google Scholar]
- 12.Haque, R., D. Mondal, P. Duggal, M. Kabir, S. Roy, B. M. Farr, R. B. Sack, and W. A. Petri, Jr. 2006. Entamoeba histolytica infection in children and protection from subsequent amebiasis. Infect. Immun. 74:904-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haque, R., D. Mondal, B. D. Kirkpatrick, S. Akther, B. M. Farr, R. B. Sack, and W. A. Petri, Jr. 2003. Epidemiologic and clinical characteristics of acute diarrhea with emphasis on Entamoeba histolytica infections in preschool children in an urban slum of Dhaka, Bangladesh. Am. J. Trop. Med. Hyg. 69:398-405. [PubMed] [Google Scholar]
- 14.Roy, S., M. Kabir, D. Mondal, I. K. M. Ali, W. A. Petri, Jr., and R. Haque. 2005. Real-time PCR assay for diagnosis of Entamoeba histolytica infection. J. Clin. Microbiol. 43:2168-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanley, S. L., Jr. 2003. Amoebiasis. Lancet 361:1025-1034. [DOI] [PubMed] [Google Scholar]
- 16.Stauffer, W., M. Abd-Alla, and J. I. Ravdin. 2006. Prevalence and incidence of Entamoeba histolytica infection in South Africa and Egypt. Arch. Med. Res. 37:266-269. [DOI] [PubMed] [Google Scholar]
- 17.Walker, E. L., and A. W. Sellards. 1913. Experimental entamoebic dysentery. Philippine J. Sci. B Trop. Med. 8:253-331. [Google Scholar]
- 18.Zaki, M., S. G. Reddy, T. F. H. G. Jackson, J. I. Ravdin, and C. G. Clark. 2003. Genotyping of Entamoeba species in South Africa: diversity, stability and transmission patterns within families. J. Infect. Dis. 187:1860-1869. [DOI] [PubMed] [Google Scholar]
- 19.Zaki, M., J. J. Verweij, and C. G. Clark. 2003. Entamoeba histolytica: direct PCR-based typing of strains using faecal DNA. Exp. Parasitol. 104:77-80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.