Abstract
Nucleic acid extraction and human immunodeficiency virus type 1 (HIV-1) genotyping using the NucliSens miniMAG platform and the TruGene HIV-1 genotyping kit gave HIV-1 sequence data from HIV-1-negative plasma spiked with 100 copies/ml reference HIV-1 RNA and from low-viremia clinical samples (<500 copies/ml) without the need for ultracentrifugation or nested second-round PCR.
Extraction of viral nucleic acid from clinical specimens is a critical step in human immunodeficiency virus type 1 (HIV-1) genotyping or any other application involving nucleic acid amplification. Failure of subsequent PCR-based applications can occur if chemical impurities or inhibitory components are carried over into the eluate or if insufficient sample is recovered due to loss or degradation of the nucleic acid during the extraction process. Consequently, good recovery of high-purity nucleic acid can allow reproducible performance of various applications with lower nucleic acid input. For example, commercially available HIV-1 genotyping or viral assay protocols typically require that clinical samples have a minimum viral level of 1,000 copies per milliliter. Antiretroviral drug resistance testing guidelines recommend that plasma samples contain at least 500 to 1,000 HIV-1 RNA copies/ml in order to achieve successful PCR amplification (3). However, genotyping assays of clinical samples with viral levels in the range of 50 to 500 copies/ml could be routinely performed in clinical testing facilities if a high-recovery, high-purity extraction process is utilized.
The NucliSens miniMAG system (bioMerieux, Inc., Durham, NC) is a semimanual extraction platform that uses magnetic silica particles and a magnetic device (6, 7). The magnetic device has a rack for 12 tubes, allowing 12 extractions to be processed at one time. Elution of nucleic acid uses the silica-based technology described by Boom et al. (1), with magnetic silica particles obviating the need for microcentrifugation during multiple washes and the final elution of nucleic acid containing both DNA and RNA molecules.
We tested the NucliSens miniMAG extraction method in combination with the TruGene HIV-1 genotyping kit (Bayer HealthCare, Tarrytown, NJ) (2, 4). HIV-1 RNA standards for testing protocol feasibility were kindly provided by the Virology Quality Assurance Laboratory (Rush-Presbyterian-St. Luke's Medical Center, Chicago, IL) as previously described (8). Clinical HIV-1 specimens were obtained from repository samples of plasma collected in sodium citrate tubes from HIV-1-positive clinical trial participants who had consented to provide samples for exploratory use. All HIV-1 plasma samples were clade B HIV-1. Viral levels in all clinical specimens had previously been determined using the COBAS Ultrasensitive v1.5 viral load assay (Roche).
To validate the protocol feasibility, we generated serial dilutions of reference HIV-1 RNA samples at 50 to 500 copies/ml as previously described (5). Serial dilutions of the Virology Quality Assurance Laboratory standards were made in Seracon I defibrinated plasma (Celliance, Atlanta, GA) to generate 32 spiked samples containing 500, 375, 100, and 50 copies/ml. One- and 2-ml plasma samples spiked with reference HIV-1 RNA were extracted using NucliSens lysis buffer and NucliSens magnetic extraction reagents (bioMerieux). The extractions were performed according to the generic protocol, with the elution buffer volume reduced to 40 μl. Briefly, 1.0- or 2.0-ml plasma samples were added to tubes containing 2.0 ml of lysis buffer. The tubes were vortexed briefly and incubated for 10 min at room temperature. Fifty microliters of magnetic silica solution was added, and the tubes were vortexed briefly. Samples were incubated for 10 min at room temperature and centrifuged for 2 min at 1,500 × g. The supernatant was removed by aspiration and the magnetic silica pellet resuspended in 400 μl of the first wash buffer and transferred to a 1.5-ml centrifuge tube. A series of washes were performed with the designated wash buffers while the tubes were in a rack on the magnetic device, with the magnet turned on to retain the magnetic silica particles as the supernatant was removed by aspiration. After the final wash, elution buffer was added, and the samples were incubated at 60°C for 5 min. Tubes were replaced in a magnetic rack, and the supernatant was transferred by pipette to a 1.5-ml centrifuge tube and stored at −80°C until use. The sequences of the protease- and reverse transcriptase-coding regions were determined using the TruGene HIV-1 genotyping sequencing kit according to the manufacturer's protocol.
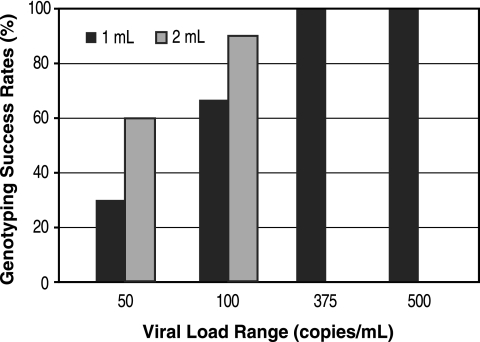
For DNA sequencing of spiked reference samples, both 1- and 2-ml input volumes were tested for samples containing 100 copies/ml (n = 12) or 50 copies/ml (n = 10). The remaining samples, containing 500 copies/ml (n = 5) or 375 copies/ml (n = 5), were extracted and tested in duplicate using 1-ml input volumes, giving a final total of 64 samples with viral levels ranging from 50 to 500 copies/ml that were subjected to sequence analysis. All the spiked HIV-1 RNA samples with >100 copies/ml were successfully sequenced using 1-ml input volumes (Fig. 1). Of the samples containing 100 copies/ml, 8 of 12 (67%) and 11 of 12 (92%) of the spiked HIV-1 RNA samples were successfully sequenced using 1- and 2-ml inputs, respectively.
FIG. 1.
Genotyping success rates with 1- and 2-ml samples (input volumes) of serial dilutions of reference HIV-1 RNA. For the samples with a viral load of 100 copies/ml, n = 12. For samples at all other concentrations, n = 10.
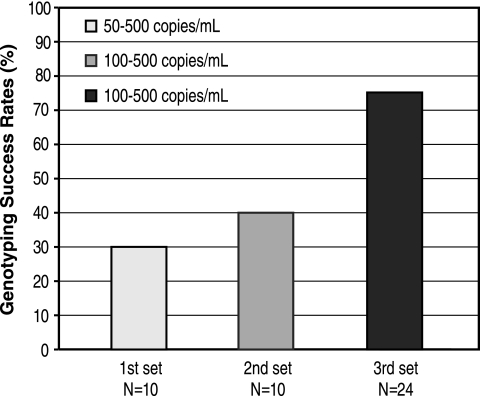
Next, we extracted a total of 44 clinical samples. Extractions of clinical samples were performed with 1-ml input volumes, using either the generic protocol (n = 20) or a slightly modified protocol (n = 24) in which the wash time was decreased to 10 seconds.
Following extractions using the generic protocol, sequence data were obtained from 7 of 20 clinical samples (35%) with viral levels ranging from 50 to 500 copies/ml. Using the modified protocol, we successfully genotyped 18 of 24 clinical samples (75%) with viral levels of between 100 and 500 copies/ml (Fig. 2). While an independent study with a larger number of samples is necessary to assess the reproducibility of our results, our preliminary findings demonstrate that low-viremia clinical samples can be successfully genotyped using this protocol.
FIG. 2.
Genotyping success rates with three sets of clinical samples. The y axis indicates the percentage of samples that were successfully sequenced.
We hypothesize that the reason why the sequencing success rate is higher with our modified extraction is probably due to less shearing and higher recovery of nucleic acid, as wash times are reduced by 66% with the modified protocol. Sequencing chromatograms showed strong nucleotide signals with low background noise (data not shown). Table 1 shows the drug resistance mutations observed in the 25 successfully genotyped clinical samples.
TABLE 1.
Drug resistance mutations observed in clinical samples (n = 25)
| Coding region | Resistance mutations
|
|
|---|---|---|
| Mutation | No. of samples | |
| Protease | L10I | 2 |
| L63P | 8 | |
| A71T | 2 | |
| None (wild-type) | 13 | |
| Reverse transcriptase | M41L | 1 |
| K103N | 2 | |
| M184V | 4 | |
| M184M/V | 1 | |
| T215D | 1 | |
| None (wild-type) | 16 | |
Appropriate management of patients with HIV-1 infection requires periodic assessment of HIV-1 viremia and monitoring of drug resistance mutations. Viral replication continues, even in patients with undetectable viral loads, and may lead to emergence of drug resistance mutations. The ability to obtain accurate genotypic data consistently from patients with low viremia (e.g., <500 to 1,000 copies/ml) would allow more effective monitoring of drug resistance.
The NucliSens miniMAG platform provides a simple, rapid, and clean extraction procedure, with no preextraction ultracentrifugation step, that yields nucleic acid of sufficient quantity and purity to allow genotyping with the TruGene HIV-1 system, even when plasma samples from individuals with low HIV-1 viremia are used. The use of the NucliSens magnetic silica particles and miniMAG takes away the need for all but one centrifugation step, and the yield and purity of the nucleic acid eluate are sufficient to allow sequencing without a nested second-step PCR. For small or mid-sized laboratories, this semimanual but convenient method of viral nucleic acid extraction and HIV-1 sequencing could result in considerable savings in costs and time.
Footnotes
Published ahead of print on 22 November 2006.
REFERENCES
- 1.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grant, R. M., D. R. Kuritzkes, V. A. Johnson, J. W. Mellors, J. L. Sullivan, R. Swanstrom, R. T. D'Aquila, M. Van Gorder, M. Holodniy, R. M. Lloyd, Jr., C. Reid, G. F. Morgan, and D. L. Winslow. 2003. Accuracy of the TRUGENE HIV-1 genotyping kit. J. Clin. Microbiol. 41:1586-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirsch, M. S., F. Brun-Vezinet, B. Clotet, B. Conway, D. R. Kritzkes, R. T. D'Aquila, L. M. Demeter, S. M. Hammer, V. A. Johnson, C. Loveday, J. W. Mellors, D. M. Jacobsen, and D. D. Richman. 2003. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society-USA panel. Clin. Infect. Dis. 37:113-128. [DOI] [PubMed] [Google Scholar]
- 4.Kuritzkes, D. R., R. M. Grant, P. Feorino, M. Griswold, M. Hoover, R. Young, S. Day, R. M. Lloyd, Jr., C. Reid, G. F. Morgan, and D. L. Winslow. 2003. Performance characteristics of the TRUGENE HIV-1 genotyping kit and the Opengene DNA sequencing system. J. Clin. Microbiol. 41:1594-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McClernon, D. R., C. Vavro, and M. St. Clair. 2006. Evaluation of a real-time nucleic acid sequence-based amplification assay using molecular beacons for detection of human immunodeficiency virus type 1. J. Clin. Microbiol. 44:2280-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rutjes, S. A., R. Italiaander, H. H. van den Berg, W. J. Lodder, and A. M. de Roda Husman. 2005. Isolation and detection of enterovirus RNA from large-volume water samples by using the NucliSens miniMAG system and real-time nucleic acid sequence-based amplification. Appl. Environ. Microbiol. 71:3734-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang, Y.-W., S. E. Sefers, H. Li, D. J. Kohn, and G. W. Procop. 2005. Comparative evaluation of three commercial systems for nucleic acid extraction from urine specimens. J. Clin. Microbiol. 43:4830-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yen-Lieberman, B., D. Brambilla, B. Jackson, J. Bremer, R. Coombs, M. Cronin, S. Herman, D. Katzenstein, S. Leung, H. J. Lin, P. Palumbo, S. Rasheed, J. Todd, M. Vahey, and P. Reichelderfer. 1996. Evaluation of a quality assurance program for quantitation of human immunodeficiency virus type 1 RNA in plasma by the AIDS Clinical Trials Group virology laboratories. J. Clin. Microbiol. 34:2695-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]