Abstract
A total of 105 multiple-antibiotic-resistant invasive pneumococcal isolates recovered in Italy from 2001 to 2003 were genetically characterized. Of these, 40 were penicillin-nonsusceptible (PNSSP) and 65 were penicillin-susceptible (PSSP) Streptococcus pneumoniae strains. Among the PNSSP isolates, 8 and 11 different restriction profiles were obtained for the pbp2b and pbp2x genes, respectively. Clonal groups were established on the basis of analysis of both pulsed-field gel electrophoresis (PFGE) types and multilocus sequence typing (MLST). Several international clones, such as Spain23F-1/ST81, Spain6B-2/ST90, Spain9V-3/ST156, and Sweden15A-25/ST263, were identified among the PNSSP isolates. Other, smaller clones, such as the minor Spanish 19F clone/ST88 and Denmark14-32/ST230, were also found. Among the PSSP isolates, clones related to England14-9/ST9, Greece6B-22/ST273, and Portugal19F-21/ST177 were found. In addition, two large clones comprised nonvaccine serotypes. One, comprising serotype 3 isolates, corresponded to the clone Netherlands3-31/ST180; the other, comprising serotype 15B/C isolates, ST474, was not related to any previously described clone. Two small clusters related to the newly described clones Greece21-30/ST193 and Netherlands15B-37/ST199 included isolates with unrelated PFGE profiles. An unusual finding was the inability to obtain the MLST allelic profile for an isolate of serotype 19A, belonging to the Sweden15A-25/ST263 clone, due to a large deletion of the xpt gene. Capsular switching was observed among both PNSSP and PSSP isolates and involved also serotypes not included in the 7-valent pneumococcal conjugate vaccine (PCV7), such as serotypes 15B/C and 19A. Since antibiotic-resistant nonvaccine serotype clones are present in Italy, continuous monitoring of pneumococcal epidemiology should be carried out in the PCV7 era.
Streptococcus pneumoniae remains a major cause of community-acquired infections worldwide. The burden of pneumococcal diseases ranges from mild respiratory tract infections to severe invasive diseases, whose associated mortality has not changed much over the last 50 years (24).
Although pneumococci can be classified into at least 90 different serotypes based on the immunochemistry of their polysaccharide capsules (17), only a limited number of serotypes are found to commonly colonize the human nasopharynx and cause disease, with major differences associated with the age of the subjects, the geographical area, and the time period under observation (16, 18, 30).
In the past decade, the emergence of pneumococcal strains resistant to antimicrobial agents has represented a major cause of concern. The appearance of penicillin-resistant pneumococci has led to the use of alternative antimicrobials for the therapy of respiratory tract infections, especially long-acting macrolides, with the result that the prevalence of both penicillin- and macrolide-resistant strains has rapidly increased worldwide (1). Surveillance of the prevalence of antibiotic-resistant pneumococcal strains over time within specific geographic areas and their genetic characterization is essential to determine the relative importance of established clones and of newly emerging ones and the impact of capsular switching. The dynamic of pneumococcal clones and serotypes is particularly important today, since the 7-valent pneumococcal conjugate vaccine (PCV7) for children has dramatically affected the epidemiology of pneumococcal diseases in the United States, where it is extensively used. The incidence of pneumococcal invasive diseases due to vaccine serotypes has decreased not only in the target pediatric population but also in the population at large (31). In parallel, however, replacement of vaccine serotypes by nonvaccine serotypes as colonizers of the nasopharynx, as well as agents of invasive disease, has been observed (19, 20). The theoretical efficacy of PCV7 in reducing the burden of invasive disease in children is lower in Europe than in North America due to a higher frequency of pediatric infections caused by the nonvaccine serotypes 1, 3, and 7F (16). In Italy, ca. 75% of invasive pneumococcal infections are estimated to be vaccine preventable in children <5 years of age (26). Although PVC7 has been available in Italy since 2001, according to a nationwide survey, in 2003 only 2.7% of children from 12 to 24 months of age had received one or more doses of the vaccine, and only 0.5% of children had completed the vaccination schedule (15).
The epidemiology of pneumococcal disease is interesting also considering the antibiotic resistance problem. In Italy, there is a discrepancy between the rate of penicillin resistance, which is moderate (ca. 10 to 12%), compared to the neighboring South European countries, and the rate of macrolide resistance (30 to 40%), which is one of the highest in the area (21, 23).
In a previous study, we described the presence of new clones of penicillin-nonsusceptible S. pneumoniae (PNSSP) in Italy and found evidence of capsular switching resulting in an unusual serotype-clone association (7).
In the present study, 105 invasive drug-resistant pneumococcus strains recovered in Italy during the period from 2001 to 2003, including 40 PNSSP and 65 penicillin-susceptible S. pneumoniae (PSSP) isolates, were characterized for their antimicrobial susceptibility patterns, restriction profiles of the penicillin-binding protein (PBP) genes, and pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) profiles in order to relate them with internationally spread clones and with clones already identified in the country.
MATERIALS AND METHODS
Pneumococcal isolates.
A total of 105 invasive pneumococcal isolates were studied. These represented all of the available (ca. 75% of the reported) multiple-drug-resistant PNSSP and PSSP strains obtained in Italy during the years from 2001 to 2003 from the nationwide surveillance of antibiotic-resistance AR-ISS (http://www.simi.iss.it/antibiotico_resistenza.htm) and the Surveillance of Bacterial Meningitis (http://www.simi.iss.it/meningite_batterica.htm).
Antibiotic susceptibility and serotyping.
Susceptibility to penicillin, ceftriaxone, erythromycin, clindamycin, tetracycline, and chloramphenicol was assayed by Etest and interpreted using the breakpoints suggested by the Clinical Laboratory Standard Institute (5). Erythromycin resistance genes were detected as previously described (23). The isolates were serotyped using the antisera produced by the Statens Serum Institute of Copenhagen, Denmark. Duplicate isolates from the same patients were excluded.
pbp2b and pbp2x gene restriction profiling and sequencing.
Restriction fragment length polymorphism of PBP genes pbp2b and pbp2x was performed as previously described (12) in all PNSSP and selected PSSP isolates. The profile designation followed that already used in past studies (7, 13). The nucleotide sequence of the transpeptidase domain of PBP2B (9) was obtained for four isolates.
PFGE and MLST.
PFGE of SmaI-restricted chromosomal DNA of S. pneumoniae isolates was performed as previously described (7). PFGE patterns were assigned designations following the type/subtype definition (13): isolates with identical profiles were assigned to the same PFGE type and subtype; isolates with similar profiles (differing by one to six bands) were assigned to different subtypes within the same PFGE type. PFGE patterns identical or similar to those found in previous studies were identified with the PFGE type and subtype designation previously used. PFGE types were analyzed with Bionumerics software for Windows (version 2.5; Applied Maths, Ghent, Belgium). Comparison was performed using the unweighted pair group method with arithmetic averages and with the Dice similarity coefficient, applying a 1.5% tolerance in band position.
MLST was performed according to the recommended method (10).
At least one isolate within each PFGE type was chosen for MLST. If different serotypes were included in a PFGE type, one isolate of each serotype was subjected to MLST. The sequence types (STs) obtained were compared to those available at the MLST database (http://www.mlst.net), with particular reference to clones recognized by the Pneumococcal Molecular Epidemiology Network (22, 28).
Nucleotide sequence accession number.
The sequence of the deleted xpt locus of strain AP033 has been assigned GenBank accession no. EF012770.
RESULTS
All of the isolates studied were obtained from sterile sites (63 from the blood and 42 from the cerebrospinal fluid) of patients admitted to 20 different hospitals throughout Italy in the years from 2001 to 2003. The age was available for 96 patients: 18 (19%) were children younger than 5 years.
Forty isolates were PNSSP, and sixty-five were PSSP. Of the 40 PNSSP isolates, 18 showed a penicillin MIC in the intermediate range (0.12 to 1 μg/ml) and 22 showed a penicillin MIC in the resistant range (2 to 4 μg/ml). Twenty-four PNSSP isolates were intermediate and one was resistant to ceftriaxone according to the breakpoints suggested for meningeal infections (5). A total of 92 isolates were resistant to erythromycin, 91 were resistant to clindamycin, 82 were resistant and 5 were intermediate to tetracycline, and 23 were resistant to chloramphenicol. All of the erythromycin-resistant isolates but one carried the erm(B) gene, conferring the MLSB phenotype. Three isolates also carried mef(E). One isolate carried mef(E) only.
pbp2b and pbp2x restriction profile analysis was performed for all PNSSP isolates and for selected PSSP isolates only, since the latter group of isolates displays low PBP variability (7, 11, 13). Two of the isolates examined did not yield any amplicon for pbp2b, whereas amplicons for pbp2x were obtained from all of the isolates. One and six new restriction profiles were obtained for pbp2b and pbp2x, respectively.
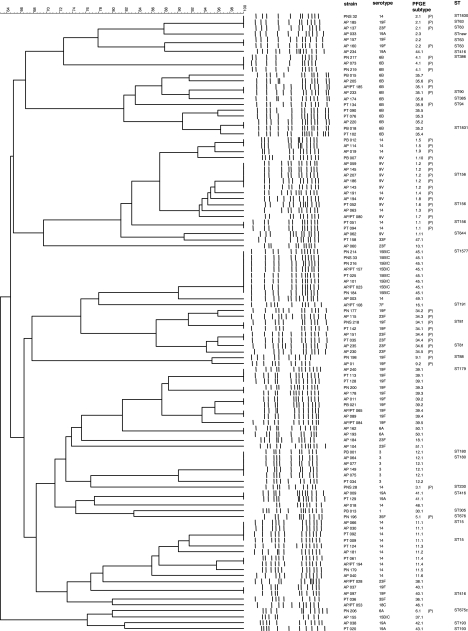
All of the isolates were genotyped by PFGE, and the dendrogram in the Fig. 1 summarizes the results. PFGE types 1 to 32 had already been described among isolates obtained in Italy in the years from 1998 to 2000 (7), although not all PFGE types previously identified were found in the present study. PFGE type 35 was designated PFGE type 1 in a study conducted on serotype 6B isolates (11). The other PFGE types were described for the first time in Italian isolates. Thirty-eight selected isolates were subjected to MLST. Clonal groups were established on the basis of analysis of both PFGE types and MLST (Table 1) . Overall, there was a good correspondence between PFGE types and MLST profiles, since isolates comprised in the same PFGE type were found to share five or more identical alleles by MLST. The only exception was represented by PFGE type 35, which comprised isolates differing by three of seven alleles.
FIG. 1.
Phylogenetic analysis of PFGE profiles obtained among the 105 S. pneumoniae isolates. The dendrogram was constructed with PFGE profiles by similarity and clustering analysis using the unweighted pair-group method with arithmetic averages and the Dice coefficient. The genetic similarity in percentages is showed above the dendrogram. Strain code, serotype, PFGE type and subtype, and ST are marked on the right. “(P)” indicates penicillin nonsusceptibility.
TABLE 1.
Clonal clusters among antibiotic-resistant invasive pneumococcal isolates in Italy
| PFGE type.subtype(s) (no. of isolates)a | Serotype(s) (no. of isolates)b | Penicillin MIC range (μg/ml) | pbp2b/pbp2x restriction pattern(s)c | Nonsusceptibility to other antibiotics (no. of isolates)d | ST (serotype examined)b | Related PMEN clone | Degree of relatedness |
|---|---|---|---|---|---|---|---|
| 1.1-1.11 (17) | 9V (9), 14 (7) | 0.25-4 | 6/2 | CX (14), EM (5), CC (5), TC (5) | 156 (9V, 14) | Spain9V-3/ST156 | IDf |
| 9V (1) | ≤0.03 | 1/1 | EM, CC, TC | 644 (9V) | DLV | ||
| 2.1-2.3 (6) | 14 (1), 19F (2), 23F (1) | 0.12-0.25 | 10/27, 10/31N, 10/35N | EM (4), CC (4), TC (4) | 63 (19F, 23F) | Sweden15A-25/ST263 | ID |
| 1830 (14) | SLV | ||||||
| 19F (1), 19A (1) | 0.03-0.06 | 10/33N, 10/34N | EM (2), CC (2), TC (2) | STNEW (19A)e | DLVg | ||
| 63 (19F) | ID | ||||||
| 3.1 (1) | 14 | 4 | 32N/32N | CX, TC | 230 | Denmark14-32/ST230 | ID |
| 4.1 (3) | 6B | 0.125-05 | 12/22 | EM, CC, TC | 386 | Poland6B-20/ST315 | DLV |
| 5.1 (1) | 35F | 0.25 | 5/2 | EM, CC, TC | 676 | None | |
| 6.1 (1) | 6A | 0.5 | 28/12 | EM, CC, TC | 675 | None | |
| 9.1-9.2 (2) | 19F | 0.12-0.25 | neg/31N | EM, CC, TC, CH | 88 | Minor Spanish 19F clone/ ST88 | ID |
| 11.1-11.6 (10) | 14 | ≤0.03 | ND | EM, CC, TC | 15 | England14-9/ST9 | SLV |
| 12.1-12.2 (6) | 3 | ≤0.03 | ND | EM (6), CC (6), TC (6), CH (4) | 180 | Netherlands3-31/ST180 | ID |
| 16.1 (1) | 7F | ≤0.03 | ND | EM, CC, TC | 191 | Netherlands7F-39/ST191 | ID |
| 30.1 (1) | 1 | 0.06 | ND | EM, CC, TC | 305 | Sweden1-40/ST304 | SLV |
| 34.1-34.6 (8) | 19F (3), 23F (5) | 2-4 | 6/2 | CX (7), EM (8), CC (8), TC (5), CH (7) | 81 (19F, 23F) | Spain23F-1/ST81 | ID |
| 35.1-35.9 (11) | 6B (4) | 0.12-2 | 22/12, 13/16 | CX (3), EM (4), CC (4), TC (4), CH (4) | 90 | Spain6B-2/ST90 | ID |
| 94 | SLV | ||||||
| 6B (7) | ≤0.03 | 1/1 | EM (7), CC (7), TC (6), CH (5) | 385 | Greece6B-22/ST273 | SLV | |
| 1831 | SLV | ||||||
| 36.1 (1) | 35F | ≤0.03 | ND | EM, CC, TC | 446 | None | |
| 37.1 (1) | 15B/C | ≤0.03 | ND | EM, CC | 474 | Greece21-30/ST193 | SLV |
| 42.1 (1) | 19A | ≤0.03 | ND | EM, CC | 193 | ID | |
| 43.1 (1) | 19A | ≤0.03 | ND | EM, CC | 193 | ID | |
| 38.1 (1) | 23F | ≤0.03 | ND | EM, CC, TC | 1832 | None | |
| 39.1-39.5 (10) | 19F | ≤0.03-0.06 | ND | EM, CC, TC | 179 | Portugal19F-21/ST177 | SLV |
| 40.1 (2) | 19F | ≤0.03 | ND | EM, CC, TC | 416 | Netherlands15B-37/ST199 | DLV |
| 41.1 (2) | 19A | ≤0.03 | ND | EM, CC, TC | 416 | DLV | |
| 44.1 (1) | 19A | 0.06 | ND | EM, CC, TC | 416 | DLV | |
| 45.1-45.3 (8) | 15B/C | ≤0.03 | ND | EM, CC, TC | 1577 | None | |
| 50.1 (2) | 6A | ≤0.03 | ND | EM, CC, TC | 1833 | None |
The following PFGE types including single PSSP isolates are not reported in the table: type 10 (serotype 23F), type 18 (serotype 23F), type 46 (serotype 18C), type 47 (serotype 33F), type 48 (serotype 14), type 49 (serotype 14), and type 51 (serotype 23F).
That is, for PFGE types comprising multiple serotypes.
A subscript “N” indicates a new profile. ND, not done.
That is, for PFGE types comprising isolates with different antibiotic susceptibilities. CX, ceftriaxone; EM, erythromycin; CC, clindamycin; TC, tetracycline; CH, chloramphenicol.
STNEW indicates that an ST number could not be assigned due to deletion of the xpt gene (see the text).
ID, identical.
Includes the xpt deletion.
In a few instances, isolates showing unrelated PFGE types, differing by >6 bands, were found to share identical MLST allelic profiles. This was seen with isolates of serotypes 19A and 19F.
Clonal groups comprising both PNSSP and PSSP strains.
Three clonal groups were shared between PNSSP and PSSP, precisely those related to the Spain9V-3, Sweden15A-25, and Spain6B-2/Greece6B-22 clones.
The largest cluster among the isolates studied was represented by the clonal group related to Spain9V-3/ST156, comprising 16 PNSSP and 1 PSSP, identified by PFGE type 1 and distributed into 11 subtypes and two serotypes, 9V and 14. Although the PNSSP isolates of both serotypes belonged to ST156, the PSSP strain belonged to ST664, a double-locus variant (DLV) of ST156. All PNSSP shared the same PBP profiles 2b-6 and 2x-2, which were previously found in isolates belonging to the Spain9V-3 clone. The PSSP isolate exhibited the wild-type profiles 2b-1 and 2x-1.
The clonal group related to Sweden15A-25/ST63 clone comprised six isolates, four PNSSP and two PSSP, characterized by PFGE type 2 and belonging to four different serotypes (14, 19A, 19F, and 23F). The identical allelic profile ST63 was shared by three PNSSP strains and one PSSP strain. The serotype 14 PNSSP showed a novel allelic combination, single-locus variant (SLV) of ST63, designed ST1830. The serotype 19A PSSP exhibited five of seven alleles that were identical to those of ST63. However, the ST of this strain could not be assigned due to the impossibility to amplify the xpt gene. Sequencing of the chromosomal region encompassing the xpt locus showed a deletion of 1,249 bp starting 776 nucleotides (nt) upstream the xpt translational start codon and including the first 473 nt of the xpt open reading frame.
Interestingly, all of the isolates belonging to this clonal group shared pbp2b profile 2b-10, whether they were PNSSP (penicillin MIC = 0.12 to 0.25 μg/ml) or PSSP (penicillin MIC = 0.03 to 0.06 μg/ml). To clarify this apparent discrepancy, the nucleotide sequence of the PBP2B transpeptidase domain (nt 1038 to 2264 according to Dowson et al. [GenBank accession no. X16022]) was obtained for two PNSSP and two PSSP isolates. The sequences of the four isolates were identical and corresponded to the PBP2B transpeptidase domain sequence of isolates with intermediate penicillin susceptibility (14, 19). Analysis of the corresponding amino acid sequence (amino acids 270 to 677) revealed 15 substitutions with respect to the sequence of R6, including the T445A mutation, considered a determinant of penicillin resistance (19). As for the pbp2x gene, two PNSSP strains shared the restriction profile 2x-27, whereas each of the other isolates showed a novel distinct profile.
A third large clonal group, comprising only serotype 6B isolates, was related to both the Spain6B-2/ST90 and the Greece6B-22/ST273 clones. In fact, these two PMEN clones differ by two alleles only by MLST and can be considered part of the same clonal complex (11). This group comprised four PNSSP and seven PSSP isolates, all characterized by PFGE type 35. The PNSSP isolates showed either an ST (ST90) identical to that of Spain6B-2 or an SLV (ST94). The PSSP showed allelic profiles (ST385 and ST1831) with three of seven alleles divergent from those of Spain6B-2, although both profiles were SLV of ST273, the allelic profile of Greece6B-22. The four PNSSP showed PBP restriction profiles typical of resistant strains, in particular the profiles 2b-22 and 2x-12, shared by two isolates, are characteristic of the clone Spain6B-2 (11).
Clonal groups comprising PNSSP only.
The most abundant clonal group comprising PNSSP only was that related to Spain23F-1/ST81. This clonal group comprised eight isolates belonging to serotypes 19F and 23F, characterized by PFGE type 34, with ST81 and PBP gene restriction profiles 2b-6 and 2x-2, which are typical of Spain23F-1. This clone, which was not found previously among Italian invasive isolates, was the only one comprising all isolates with full penicillin resistance (penicillin MIC = 2 to 4 μg/ml) (7).
Two small clonal groups were also identified. One was related to Poland6B-20/ST315 and comprised three serotype 6B isolates, characterized by PFGE type 4, PBP restriction profiles 2b-12 and 2x-22 (11), and ST386, a DLV of ST315.
The other clonal group comprised two serotype 19F isolates and was characterized by PFGE type 9 and ST88, which in the MLST database correspond to the “minor Spanish multiresistant serotype 19F clone.” These two isolates did not yield any PCR amplicon for pbp2b, a characteristic already observed in this clonal group (7).
The remaining three PNSSP isolates showed distinct PFGE types and MLST profiles that had been already identified in previous studies, suggesting that they represent minor PNSSP clones that are established in Italy. A serotype 14 isolate, PFGE type 3, ST230, belongs to the recently described Denmark14-32 clone, which was previously found in serotype 24 PNSSP isolates from Naples (27). A serotype 35F isolate shared PFGE type 5 and ST676 with serotype 35F PNSSP isolates previously described. Similarly, a serotype 6A isolate, shared PFGE type 6 and ST675 with a previously characterized serotype 6A PNSSP (7).
Clonal groups comprising PSSP only.
Two large clonal groups had been already described among Italian PSSP isolates. One, related to England14-9/ST9, comprised 10 serotype 14 isolates showing the allelic profile ST15, an SLV of ST9. This was the only clonal group that comprised a consistent number of isolates (5 of 10) obtained from children <5 years of age (data not shown). The other clonal group corresponded to Netherlands3-31/ST180 and comprised six serotype 3 isolates (7).
Single isolates belonged to clonal groups already encountered in previous studies: a serotype 7F isolate, PFGE type 16, was related to The Netherlands7F-39/ST191 clone, and a serotype 1 isolate, PFGE type 30, was related to the clone Sweden1-40/ST304.
Two new clusters comprising multiple isolates were identified. One cluster was related to Portugal19F-21/ST177 and comprised 10 serotype 19F isolates characterized by PFGE type 39 and ST179, an SLV of ST177. The other cluster included eight serotype 15B/C isolates, PFGE type 45, and ST1577. In the MLST database, ST1577 is only associated with a single serotype 15A isolate, and there are only two other strains that are DLVs of ST1577.
Two serotype 6A isolates, PFGE type 50, belonged to the novel allelic profile ST1833. In the MLST database, several serogroup 6 isolates are reported that are SLV or DLVs of ST1833.
A serotype 35F isolate, PFGE type 36, yielded ST446, shared with another serotype 35F isolate obtained in the Oxford carriage study (3). A 23F isolate, PFGE type 38, showed the novel profile ST1832. In the MLST database, various serotype 23F isolates were present that were SLVs or DLVs of ST1832.
Two additional clonal groups were observed that enclosed isolates that were unrelated according to PFGE criteria, since their profiles differed by >6 bands. One clonal group, related to the newly described clone Greece21-30/ST193, included two serotype 19A isolates sharing ST193 but showing different PFGE types (types 42 and 43, respectively) and a serotype 15B/C isolate, PFGE type 37, ST474, an SLV of ST193. The other clonal group, related to another newly described clone, Netherlands15B-37/ST199, comprised five isolates, two serotype 19F isolates, PFGE type 40, and three serotype 19A isolates belonging to PFGE types 41 and 44. All of these isolates shared ST416, a DLV of ST199.
DISCUSSION
In the past two decades, increased antibiotic use has been the most important selective force driving the appearance and circulation of new pneumococcal clones (1, 2). Today, the epidemiology of S. pneumoniae disease is undergoing profound changes in the countries where PCV7 has been amply introduced, both in the target pediatric population and in older individuals (31).
Due to the limited use of PCV7 in Italy in the years from 2001 to 2003, the pneumococcal epidemiology described here can be considered to reflect a prevaccine situation.
Until recently, most genotyping studies were circumscribed to PNSSP, conveying the false impression that clonality was associated with penicillin resistance. In fact, studies directed at the pneumococcal population in general have shown a dynamic based on the circulation of particular clones among PSSP as well (4, 11, 18). The isolates examined in our study included not only PNSSP isolates but also PSSP isolates that were multidrug resistant. The majority of the isolates examined were resistant to erythromycin, clindamycin, and tetracycline. Resistance to chloramphenicol was limited to a few clones of both PNSSP isolates, such as Spain23F-1, and PSSP isolates, such as Greece6B-22, Netherlands3-31, and the minor Spanish multiresistant 19F clone.
We confirmed the presence of PNSSP clones already detected in Italy in past studies, such as the internationally spread PMEN clones Spain23F-1/ST81, Spain6B-2/ST90, Spain9V-3/ST156, and Sweden15A-25/ST263. Clone Spain23F-1 was not detected in our previous study of invasive isolates (7), although it had been described in Italian isolates from 1996 to 1997 (21). This clone is the only one comprising isolates with penicillin MICs of ≥2 μg/ml, and its expansion is likely the cause of an increase in the percentage of pneumococci with full penicillin resistance in Italy in the last years (data not shown). In addition, most isolates belonging to this clone are also not susceptible to ceftriaxone and chloramphenicol, indicating that this is the most drug-resistant pneumococcal clone circulating in Italy.
Among the PSSP clones, other than the already described clones related to England14-9/ST9 and Greece6B-22/ST273 (6, 11, 23), we found a serotype 19F clone corresponding to Portugal19F-21/ST177 and two large clones comprising nonvaccine serotypes. One, related to Netherlands3-31 clone/ST180, enclosed serotype 3 isolates, the other was unrelated to previously described clones and comprised isolates belonging to serotype 15B/C.
The phenomenon of capsular switching was not limited to isolates within PNSSP international clones, where this occurrence has been amply described (22), but was also noted among PSSP isolates. Both vaccine and nonvaccine serotypes, such as serotypes 14, 9V, 15B/C, 19A, 19F, and 23F, were involved in capsular switching. Capsular switching among nonvaccine serotypes has been reported to be increasingly frequent (25). For example, the clonal cluster related to Sweden15A-25, comprised four different serotypes other than 15A. This same clonal cluster comprised isolates with unusual features, such as two PSSP with a PBP2B transpeptidase domain sequence identical to that of the PNSSP of the group. One of these PSSP isolates bore a large nucleotide deletion in the xpt locus that hindered amplification of the xpt gene for the MLST scheme. This finding is very unusual, since MLST is based on the allelic sequences of housekeeping genes, which are considered essential. However, in Bacillus subtilis the xpt gene that encodes a xanthine phosphoribosyltransferase can be disrupted without causing any major phenotype (29). In pneumococcus, xpt allele 113 has a large internal in-frame deletion. Our finding confirms that large deletions involving xpt can occur that may lead to difficulties in applying the MLST scheme or analyzing the MLST data (8).
Among PSSP strains, the majority of the clonal groups comprised isolates belonging to the same serotype. In some cases, this correspondence was limited to isolates circulating in our country because the MLST database usually also contains different serotypes. For example, clonal group ST1577, comprising only serotype 15B/C isolates, shares identical or related STs with isolates belonging to serotypes 15A and 6B from other countries. However, we have observed few examples of capsular switching among PSSP isolates of serotype 19A, 19F, and 15B of the clonal groups Netherlands15B-37/ST199 and Greece21-30/ST193. Interestingly, in the MLST database, isolates of the latter clone belong to at least eight different serotypes.
The circulation of clonal clusters of multidrug-resistant pneumococci of nonvaccine serotypes is particularly threatening. Although serotypes 1, 3, and 19A will be included in the new 13-valent conjugate vaccine, serogroup 15 remains untargeted. Drug-resistant isolates of serogroup 15 are on the rise in the United States following the introduction of PCV7 and are a cause for concern (14).
Recently, in several Italian regions, implementation of the pneumococcal vaccination has been carried out in the pediatric population. Therefore, the spreading of nonvaccine clones is predicted to further increase, and continuous surveillance will be important to follow future changes.
Acknowledgments
We thank Elisa Franchin for skilled technical assistance and Monica Monaco for serotyping the isolates.
This study was supported in part by grants from Regione Veneto (RSF 168/04 and RSF 61/02) and from the Italian Ministero della Salute (Progetto Finalizzato 2003 Controllo delle Infezioni Respiratorie and Progetto Sorveglianza della Resistenza agli Agenti Antimicrobici 2002).
Footnotes
Published ahead of print on 22 November 2006.
REFERENCES
- 1.Albrich, W., D. Monnet, and S. Harbarth. 2004. Antibiotic selection pressure and resistance in Streptococcus pneumoniae and Streptococcus pyogenes. Emerg. Infect. Dis. 10:514-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boccia, D., S. Spila Alegiani, A. Pantosti, M. L. Moro, and G. Traversa. 2004. The geographic relationship between the use of antimicrobial drugs and the pattern of resistance for Streptococcus pneumoniae in Italy. Eur. J. Pharmacol. 60:115-119. [DOI] [PubMed] [Google Scholar]
- 3.Brueggemann, A., D. Griffiths, E. Meats, T. Peto, D. Crook, and B. Spratt. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187:1424-1432. [DOI] [PubMed] [Google Scholar]
- 4.Camilli, R., E. Pettini, M. Del Grosso, G. Pozzi, A. Pantosti, and M. Oggioni. 2006. Zinc metalloproteinase genes in clinical isolates of Streptococcus pneumoniae: association of the full array with a clonal cluster comprising serotypes 8 and 11A. Microbiology 152:313-321. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement. M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Del Grosso, M., F. Iannelli, C. Messina, M. Santagati, N. Petrosillo, S. Stefani, G. Pozzi, and A. Pantosti. 2002. Macrolide efflux genes mef(A) and mef(E) are carried by different genetic elements in Streptococcus pneumoniae. J. Clin. Microbiol. 40:774-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dicuonzo, G., G. Gherardi, R. E. Gertz, F. D'Ambrosio, A. Goglio, G. Lorino, S. Recchia, A. Pantosti, and B. Beall. 2002. Genotypes of recent invasive pneumococcal isolates recovered from Italian patients. J. Clin. Microbiol. 40:3660-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diggle, M. A., and S. C. Clarke. 2005. Truncated xpt gene present in invasive Streptococcus pneumoniae may have implications for MLST schemes. J. Med. Microbiol. 54:909-912. [DOI] [PubMed] [Google Scholar]
- 9.Dowson, C. G., A. Hutchison, J. A. Brannigan, R. C. George, D. Hansman, J. Linares, A. Tomasz, J. M. Smith, and B. G. Spratt. 1989. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 86:8842-8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 11.Gherardi, G., M. Del Grosso, A. Scotto d'Abusco, F. D'Ambrosio, G. Dicuonzo, and A. Pantosti. 2003. Phenotypic and genotypic characterization of two penicillin-susceptible serotype 6B Streptococcus pneumoniae clones circulating in Italy. J. Clin. Microbiol. 41:2855-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gherardi, G., J. S. Inostrozo, M. O'Ryan, V. Prado, S. Prieto, C. Arellano, R. R. Facklam, and B. Beall. 1999. Genotypic survey of recent beta-lactam-resistant pneumococcal nasopharyngeal isolates from asymptomatic children in Chile. J. Clin. Microbiol. 37:3725-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gherardi, G., C. G. Whitney, R. R. Facklam, and B. Beall. 2000. Major related sets of antibiotic-resistant pneumococci in the United States as determined by pulsed-field gel electrophoresis and pbp1a-pbp2b-pbp2x-dhf restriction profiles. J. Infect. Dis. 181:216-229. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez, B., K. Hulten, L. Lamberth, S. Kaplan, E. J. Mason, and the U.S. Pediatric Multicenter Pneumococcal Surveillance Group. 2006. Streptococcus pneumoniae serogroups 15 and 33: an increasing cause of pneumococcal infections in children in the United States after the introduction of the pneumococcal 7-valent conjugate vaccine. Pediatr. Infect. Dis. J. 25:301-305. [DOI] [PubMed] [Google Scholar]
- 15.Gruppo di Lavoro ICONA. 2003. ICONA 2003: indagine nazionale sulla copertura vaccinale infantile. Rapporti ISTISAN 03/37. Istituto Superiore di Sanità, Rome, Italy.
- 16.Hausdorff, W. P., J. Bryant, P. R. Paradiso, and G. R. Siber. 2000. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use, part I. Clin. Infect. Dis. 30:100-121. [DOI] [PubMed] [Google Scholar]
- 17.Henrichsen, J. 1995. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 33:2759-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henriques Normark, B., M. Kalin, A. Ortqvist, T. Akerlund, B. Liljequist, J. Hedlund, S. Svenson, J. Zhou, B. Spratt, S. Normark, and G. Kallenius. 2001. Dynamics of penicillin-susceptible clones in invasive pneumococcal disease. J. Infect. Dis. 184:861-869. [DOI] [PubMed] [Google Scholar]
- 19.Huang, S., R. Platt, S. Rifas-Shiman, S. Pelton, D. Goldmann, and J. Finkelstein. 2005. Post-PCV7 changes in colonizing pneumococcal serotypes in 16 Massachusetts communities, 2001 and 2004. Pediatrics 116:408-413. [DOI] [PubMed] [Google Scholar]
- 20.Kyaw, M., R. Lynfield, W. Schaffner, A. Craig, J. Hadler, A. Reingold, A. Thomas, L. Harrison, N. Bennett, M. Farley, R. Facklam, J. Jorgensen, J. Besser, E. Zell, A. Schuchat, C. Whitney, and the Active Bacterial Core Surveillance of the Emerging Infections Program Network. 2006. Effect of introduction of the pneumococcal conjugate vaccine on drug-resistant Streptococcus pneumoniae. N. Engl. J. Med. 354:1455-1463. [DOI] [PubMed] [Google Scholar]
- 21.Marchese, A., L. Gualco, I. Cochetti, M. Montanari, A. Speciale, S. Musumeci, P. Varaldo, G. Nicoletti, and G. Schito. 2005. Antibiotic susceptibility and serotype distribution in Streptococcus pneumoniae circulating in Italy: results of the SEMPRE surveillance study (2000-2002). Int. J. Antimicrob. Agents 26:138-145. [DOI] [PubMed] [Google Scholar]
- 22.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monaco, M., R. Camilli, F. D'Ambrosio, M. Del Grosso, and A. Pantosti. 2005. Evolution of erythromycin resistance in Streptococcus pneumoniae in Italy. J. Antimicrob. Chemother. 55:256-259. [DOI] [PubMed] [Google Scholar]
- 24.Obaro, S. K., M. A. Monteil, and D. C. Henderson. 1996. Fortnigtly review: the pneumococcal problem. BMJ 312:1521-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pai, R., M. Moore, T. Pilishvili, R. Gertz, C. Whitney, B. Beall, and the Active Bacterial Core Surveillance Team. 2005. Postvaccine genetic structure of Streptococcus pneumoniae serotype 19A from children in the United States. J. Infect. Dis. 192:1988-1995. [DOI] [PubMed] [Google Scholar]
- 26.Pantosti, A., D. Boccia, F. D'Ambrosio, S. Recchia, G. Orefici, M. Moro, the National Surveillance of Bacterial Meningitis, and the EARSS-Italia groups. 2003. Inferring the potential success of pneumococcal vaccination in Italy: serotypes and antibiotic resistance of isolates from invasive diseases. Microb. Drug Resist. 9:S61-S68. [DOI] [PubMed] [Google Scholar]
- 27.Pantosti, A., G. Gherardi, M. Conte, F. Faella, G. Dicuonzo, and B. Beall. 2002. A novel, multiple drug-resistant, serotype 24F strain of Streptococcus pneumoniae that caused meningitis in patients in Naples, Italy. Clin. Infect. Dis. 35:205-208. [DOI] [PubMed] [Google Scholar]
- 28.Pneumococcal Molecular Epidemiology Network. 15. October 2006, posting date. [Online.] http://www.sph.emory.edu/PMEN/pmen_clone_collection.html. PMEN, Hubert Department of Global Health, Rollins School of Public Health, Emory University, Atlanta, GA.
- 29.Saxild, H. H., and P. Nygaard. 1987. Genetic and physiological characterization of Bacillus subtilis mutants resistant to purine analogs. J. Bacteriol. 169:2977-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott, J. A. G., A. J. Hall, R. Dagan, J. M. S. Dixon, S. J. Eykyn, A. Fenoll, M. Hortal, L. P. Jetté, J. H. Jorgensen, F. Lamothe, C. Latorre, J. T. Macfarlane, D. M. Shlaes, L. E. Smart, and A. Taunay. 1996. Serogroup-specific epidemiology of Streptococcus pneumoniae: association with age, sex, and geography in 7,000 episodes of invasive disease. Clin. Infect. Dis. 22:973-981. [DOI] [PubMed] [Google Scholar]
- 31.Whitney, C. G., M. Farley, J. Hadler, L. Harrison, N. Bennett, R. Lynfield, A. Reingold, P. Cieslak, T. Pilishvili, D. Jackson, R. R. Facklam, J. H. Jorgensen, A. Schuchat, and the Active Bacterial Core Surveillance of the Emerging Infections Program Network. 2003. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348:1737-1746. [DOI] [PubMed] [Google Scholar]