Abstract
Rotavirus infections by G12 strains in several countries have recently been described. In this study, we report the emergence of G12 strains in south India. Fourteen cases of G12 infection were identified between June and September 2005. G12 was seen in combination with P[6], P[8], or nontypeable P type. Nine cases, including five symptomatic infections and four asymptomatic infections, were identified as part of routine surveillance for rotavirus infections in a birth cohort in the community between June and July 2005. Significant temporal and time-space clustering of eight of these cases represents a possible recent introduction of a new rotavirus VP7 genotype. Previous rotavirus infections had been documented for six of the nine children in the community. In the following 2 months, five cases of G12 infection were identified among children presenting to a referral hospital with diarrhea. This is the first description of symptomatic and asymptomatic G12 infections in children in the community. The detection of G12 strains from different parts of the world in recent years suggests the possibility of its emergence as an important global genotype. Monitoring of cocirculating rotavirus strains and detection of emerging strains is important in the context of the availability of rotavirus vaccines.
Rotaviruses are the major cause of severe gastroenteritis in infants and young children worldwide. It is estimated that rotavirus is responsible for 611,000 deaths annually, with 80% of these taking place in poorer countries (19). Severe, life-threatening disease in children is caused predominantly by group A rotaviruses. Vaccines currently appear to offer the most promising tool for preventing morbidity and mortality caused by rotavirus.
Rotaviruses are double-stranded RNA viruses comprising a genus within the family Reoviridae. The genome consists of 11 segments of double-stranded RNA that code for six structural viral proteins (VP1, VP2, VP3, VP4, VP6, and VP7) and six nonstructural proteins (NSP1 to NSP6) (7). Rotaviruses are classified into seven different groups (A to G) based on the antigenic specificity of the VP6 capsid proteins. Variability in the genes encoding the outer capsid proteins VP7 and VP4 forms the basis of the current strain typing of group A rotaviruses into G and P genotypes, respectively. At least 15 G genotypes and 26 P genotypes have been identified to date (15).
The epidemiology of rotavirus is complex and varies in different settings. The use of molecular methods has provided increased sensitivity for detection and characterization of unusual strains apart from commonly described genotypes. The G12 strain was first identified and serologically characterized in 1990 from children with gastroenteritis in the Philippines. The strain was not observed again until 2002, when one case each was detected in Thailand (21, 26) and the United States (10). In the following years, more cases of G12 rotavirus infection were described from India (6), Japan (25), Korea (24), and Argentina (3). To date, 45 cases of human G12 rotavirus infection have been described worldwide. Of these, 28 cases were reported among children of <4 years of age in eastern India, including a recent report wherein G12 rotaviruses were found to be responsible for 25 of 147 rotavirus infections (17%), making it the third most common genotype, after G1 and G2, in that area (23).
We have been carrying out concurrent surveillance programs for rotavirus infection and disease in the community and among hospitalized children in Vellore, India, which is in the southern part of the country (1). In this study, we describe a possible outbreak of G12 rotavirus infection in the community and the subsequent detection of the genotype among hospitalized children.
MATERIALS AND METHODS
Rotavirus surveillance in the community.
A cohort of 452 children were recruited consecutively from March 2002 to August 2003 from an urban slum area in Vellore, India. The study area was mapped between November 2001 and August 2002, and pregnant women were identified through repeated household surveys and from local antenatal clinics. Sociodemographic, environmental, and health care-related characteristics were collected at baseline. Fecal samples were collected from all children fortnightly as part of routine surveillance for rotavirus infection by a field worker who enquired about diarrhea and other morbidities and examined the children. Additionally, fecal samples were collected from any child who developed diarrhea, identified either by a routine field worker visit or by self-referral by the mother. Stool samples were collected every 2 days until the cessation of diarrhea.
Hospital-based surveillance.
Rotavirus surveillance among children hospitalized with diarrhea was carried out at the Christian Medical College, a 1,800-bed referral hospital in Vellore, India. The number of children under 5 years of age in Vellore is estimated to be approximately 42,000. Children requiring hospitalization for diarrhea were usually residents of the town or of neighboring areas. Stool samples were collected from all children under 5 years of age presenting with acute gastroenteritis and requiring hospitalization for at least 6 h for rehydration. Additionally, stool samples were collected from older children with acute gastroenteritis in whom a viral causative agent was suspected by the attending physician.
Assessment of severity.
Careful scoring of clinical data was carried out for all children. Diarrhea was defined as passage of three or more watery stools in a 24-h period. The severity of diarrhea was assessed with the Vesikari scoring system (22), based on the duration of diarrhea, maximum number of stools passed per day, duration and peak frequency of vomiting, degree of fever, presence and severity of dehydration, and treatment. For calculations of the severity scores for children in the community, a modified Vesikari scoring system was followed. Since accurate temperature measurements were not possible in the field, temperatures were recorded as normal, low-grade fever, and high-grade fever, as reported by caregivers. An episode was considered mild for a score of ≤5, moderately severe for a score of 6 to 10, and severe for a score of >10.
Laboratory methods.
All samples were screened for rotavirus with an enzyme immunoassay for detection of VP6 antigen (Rota IDEIA; Dako Ltd., Denmark) according to the manufacturer's instructions. Viral RNA was extracted from rotavirus-positive fecal suspensions by the guanidine isothiocyanate-silica method described by Boom et al. (2), followed by cDNA synthesis with random primers [pd(N)6 hexamers; Pharmacia Biotech, United Kingdom] and Moloney murine leukemia virus reverse transcriptase (Invitrogen/Life Technologies, United Kingdom). The cDNA was used as a template for G- and P-typing heminested multiplex PCRs using published oligonucleotide primers and methods (8, 9, 11, 12). Purified first-round PCR amplicons for samples for which a G genotype could not be assigned but an 881-bp first-round product was detected were taken for cycle sequencing with fluorescent dideoxy chain terminators (Big Dye terminator cycle sequencing reagent set; Applied Biosystems) and sequenced with an automated genetic analyzer (ABI 310 analyzer; PE Applied Biosystems, Foster City, CA).
Statistical methods.
The incidence rates of G12 infection during consecutive time periods were calculated as the number of infections per person time at risk. Age at first infection of G12 was compared to infection with other G types by the Mann-Whitney test. All analyses were done with STATA, version 8.0 (Stata Corp., College Station, TX).
Spatial analysis was performed to determine the incidence of G12 rotavirus infections from birth until March 2006, to detect any clustering of episodes, with SaTScan, version 6.0 (17). The streets and study houses of the urban slums had been mapped and georeferenced previously with ArcView GIS 3.3 (Environmental Systems Research Inc.). The waypoints and trackpoints were collected with GPS Garmin V and downloaded as layers by GPS Utility, version 4.10.4. (GPS Utility Ltd., Southampton, England). Identification of space-time clusters for high rates by the Poisson probability model (16), with a maximum spatial cluster size of 25% of the total population, was done with the equation
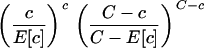 |
where C is the total number of cases, c is the observed number of cases within the window, and E[c] is the covariate-adjusted expected number of cases within the window under the null hypothesis. Significance (P value) was evaluated with the Monte Carlo simulation.
RESULTS
G12 rotavirus infections.
Sequencing of first-round amplicons of untyped G types revealed that they were G12 strains. Fourteen cases of G12 rotavirus infection were identified, with nine cases from the community and five in hospitalized children. Among the G12 cases identified from the community, five samples were collected during diarrheal episodes and four samples were from asymptomatic children collected during routine surveillance. The median severity scores of diarrhea among symptomatic children in the community and hospital were 8 and 11, respectively. The time points of detection of G12 cases and the clinical and demographic characteristics are summarized in Table 1.
TABLE 1.
Clinical and demographic features of G12 cases identified in Vellore, India
| Patient | Sample identification no.a | Date of infection | Location | Symptoms | Age (mo) | Sex | Vesikari score | G type | P typeb |
|---|---|---|---|---|---|---|---|---|---|
| 1 | CRI 32215 | 22 June 2005 | Community | Asymptomatic | 23 | Male | G12 | P[6] | |
| 2 | CRI 32442 | 26 June 2005 | Community | Symptomatic | 24 | Male | 4 | G12 | UT |
| 3 | CRI 32450 | 30 June 2005 | Community | Asymptomatic | 29 | Male | G12 | P[6] | |
| 4 | CRI 32548 | 2 July 2005 | Community | Symptomatic | 38 | Female | 8 | G12 | P[6] |
| 5 | CRI 32608 | 6 July 2005 | Community | Symptomatic | 29 | Female | 18 | G12 | P[6] |
| 6 | CRI 32609 | 6 July 2005 | Community | Symptomatic | 28 | Female | 13 | G12 | P[8] |
| 7 | CRI 32742 | 6 July 2005 | Community | Symptomatic | 23 | Male | 6 | G12 | P[8] |
| 8 | CRI 32920 | 13 July 2005 | Community | Asymptomatic | 36 | Female | G12 | P[6] | |
| 9 | CRI 33015 | 20 July 2005 | Community | Asymptomatic | 32 | Female | G12 | P[6] | |
| 10 | RV 403 | 3 August 2005 | Hospital | Symptomatic | 13 | Female | 12 | G12 | P[6] |
| 11 | RV 412 | 20 August 2005 | Hospital | Symptomatic | 120 | Female | 11 | G12 | P[8] |
| 12 | RV 413 | 21 August 2005 | Hospital | Symptomatic | 12 | Male | 13 | G12 | P[6] |
| 13 | RV 417 | 23 August 2005 | Hospital | Symptomatic | 4 | Female | 6 | G12 | P[6] |
| 14 | RV 430 | 22 September 2005 | Hospital | Symptomatic | 11 | Male | 10 | G12 | P[6] |
The community surveillance was 1,270 child years of follow-up from March 2002 to March 2006. Of the 452 children, 328 were still being monitored as of March 2006. During this 4-year follow-up period, 282 children were found to have 532 rotavirus infections, including both symptomatic and asymptomatic infections. The different genotypes detected were G1 (n = 155), G2 (n = 91), G3 (n = 1), G4 (n = 2), G8 (n = 1), G9 (n = 59), G10 (n = 69), G12 (n = 9), and mixed G types (n = 31). One hundred fourteen samples could not be G typed. No first-round PCR product could be detected for 101 of these samples, and all 13 samples that had a first-round product were G12. An additional G12 strain was identified by screening of untyped samples with a modified PCR incorporating a newly designed G12 primer (I. Banerjee et al., unpublished data). The age at first infection with a G12 strain was compared with the age at first infection for other G genotypes in the community and was found to be significantly higher than the common G types in this setting (Table 2). The overall median age of infection for G12 rotavirus in the community (28.5 months; interquartile range [IQR], 23.8 to 32.1 months) was also higher than the median age of G12 infection among hospitalized children (12 months; IQR, 11 to 13 months) (P = 0.08 [Mann-Whitney test]).
TABLE 2.
Comparison of age at first infection of rotavirus G12 with other G-type rotavirus infections detected by multiplex PCR in a community-based birth cohorta
| G type | nb | Median age (mo) at first infection (IQR) | Pc |
|---|---|---|---|
| G1 | 114 | 7.9 (4.6-17.1) | <0.001 |
| G2 | 83 | 13.9 (8.9-18.1) | <0.001 |
| G9 | 52 | 22.7 (11.7-28.5) | 0.02 |
| G10 | 65 | 0.5 (0.3-2.5) | <0.001 |
| Nontypeable | 83 | 11.9 (3.5-20.5) | <0.001 |
| Rare (G3, G4, G8) | 4 | 13.7 (8.5-24.5) | 0.06 |
| Mixed | 30 | 25.4 (11.5-30.9) | 0.21 |
| G12 | 9 | 28.49 (23.77-32.07) |
A community-based birth cohort of 452 children was recruited over a period of 18 months, from March 2002 to August 2003. As of March 2006, 328 children were still under follow-up. During this 4-year follow-up period, 282 children had 542 rotavirus infections. The age at first infection with genotype G12 was compared with that for other G genotypes.
n, number of children infected with the indicated G type.
P values were determined with the Mann-Whitney test by comparing G-type-specific medians to those for G12.
Association of G12 with different P types.
Of the fourteen G12 rotavirus infections identified in Vellore, 13 strains were associated with either P type P[6] or P[8]. These include 10 cases of G12P[6] infection (community, 6; hospital, 4) and 3 cases of G12P[8] infection (community, 2; hospital, 1). The P type could not be determined for one sample from the community. All asymptomatic infections in the community were caused by the G12P[6] strain.
Calendar time distribution of G12 detection.
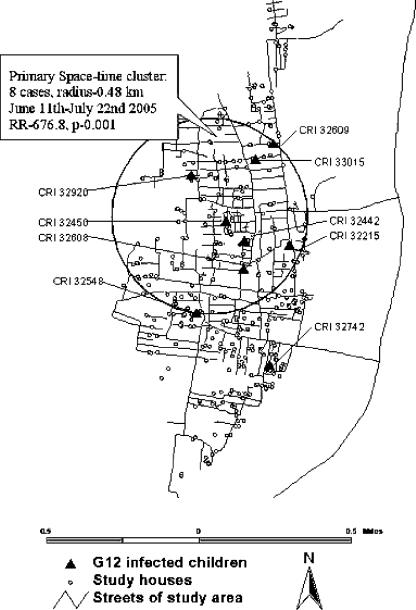
During the initial 972 child years of surveillance (March 2002 to May 2005), no G12 strains were detected. The first G12 strain was detected on 22 June 2005. The rate of G12 infection was 12.77 (95% confidence interval, 6.75 to 24.55) per 100 child months during June and July 2005. All strains from the community were identified between 22 June and 20 July 2005 from children who lived in adjacent neighborhoods, possibly representing an outbreak with a genotype not detected previously in this setting. Spatial analysis of G12 cases from the community showed a significant space-time cluster involving eight cases within an area of 0.48 km radius during the period of 11 June to 22 July 2005 (relative risk, 676.8; P, 0.001) (Fig. 1). No further G12 strains were detected from the community during surveillance carried out in the following 8 months (August 2005 to March 2006 [292 child years of follow-up]).
FIG. 1.
Space-time clustering of G12 strains in the community, showing the primary cluster of G12 infections during the period of June and July 2005. The circled area is the geographic base of the cylinder (11 June to 22 July), encompassing eight cases of infection; this was determined to be the cluster with the most likely and significant relative risk of infection (RR), 676.8 (within versus outside the cluster).
The five cases of G12 infection among hospitalized patients were seen in the two months (July and August 2005) following identification of rotavirus G12 in the community. The hospitalized children did not appear to be clustered in any particular area; they were from different areas in and around Vellore.
History of previous rotavirus infection/disease.
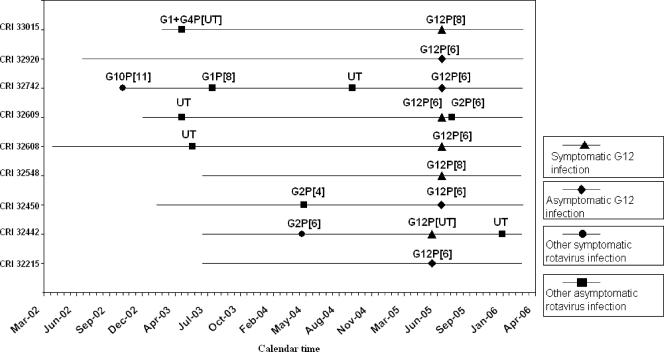
There were no records of previous rotavirus infection in hospitalized children, because rotavirus is not routinely tested in children with diarrhea presenting to hospital; however, records of children in the birth cohort were available. Of the five symptomatic children with G12 infection in the community, four had previously documented rotavirus infections. Symptomatic rotavirus infection with the G2P[6] strain was seen in one child, while the other three children previously had asymptomatic rotavirus infections (one G1/G4 strain and two nontypeable strains). Of the four children with asymptomatic G12 infection, two had no previous rotavirus infections, one had an asymptomatic G2P[4] infection, and one had three prior rotavirus infections: a symptomatic infection with the G10P[11] strain and two asymptomatic infections with a G1P[8] strain and a nontypeable strain. The period between previous rotavirus infections and infection with a G12 strain ranged from 9 to 25 months (Fig. 2).
FIG. 2.
Calendar time of rotavirus infections with G12 and other genotypes for the nine children in the community. Each line represents follow-up of a child from recruitment until March 2006. The points at which rotavirus infections were seen are plotted for each child. All G12 strains in the community were identified during June and July 2005. UT, untyped.
DISCUSSION
In this report, we describe the first detection of G12 rotaviruses in south India. Nine cases of G12 rotavirus infection were detected in the community in June and July 2005. The highly significant temporal and time-space cluster in the community is likely to represent an outbreak of a new VP7 genotype of rotavirus in the community. In the following 2 months, five cases of rotavirus infection with G12 strains were identified among patients presenting to a referral hospital in Vellore. The patients did not appear to be from any particular area and were younger than the G12-infected children in the community.
Introduction of a new strain of rotavirus in an urban slum setting presents a potentially high probability of transmission of infection. The slums are overcrowded, with closely clustered houses, many rubbish dumps, and open drains. Sewage and drinking water sources are often close to each other. Defecation close to water sources also increases the possibility of spread of enteric pathogens.
The median Vesikari severity scores of the symptomatic children in the community and hospital were 8 and 11, respectively. These are similar to the severity of rotavirus infection with other G types described previously from the same setting (1). The difference in Vesikari scores among the two groups may be due to an inherent referral bias, as more-severe cases require admission to the hospital. Also, close monitoring of the children in the community children may result in the earlier identification of symptomatic children and prompt treatment. In this study, four cases of asymptomatic infection by G12P[6] were also identified. This is the first report of asymptomatic infection with G12 rotavirus.
The most recent previous study of the molecular epidemiology of rotavirus diarrhea in this setting showed that in 2003 and 2004, P[8] was seen among 15.8% and 55.2% of all rotavirus diarrhea in the community and hospital, respectively. The P[6] genotype was infrequent, detected in 1.2% and 2.2% of infections in the community and hospital, respectively (1). During the period of June to August 2005, P[6] and P[8] were the most prevalent P genotypes seen in both the community and hospital. Seven cases each of P[6] and P[8] infection were seen in the community of 23 rotavirus-positive samples detected in this period. P[8] was seen in combination with G1 (n = 1), G9 (n = 1), G12 (n = 2), and G1/G2 (n = 1), while two samples could not be G typed. P[6] was seen with G genotypes G12 (n = 6) and G2 (n = 1). Of 11 rotavirus-positive samples among hospitalized patients during the same period, four cases each of P[6] and P[8] infection were seen, while three were P[6]/P[8] mixed infections. All four P[6] strains in hospitalized patients were seen in combination with G type G12. The only other P[6] infection was an asymptomatic G2P[6] infection seen soon after a symptomatic G12P[6] infection in a child in the community cohort (Fig. 2). In a setting where P[6] strains have not been frequently described in the past, the detection of a G12P[6] strain may be part of the emergence of a new strain. The less frequent association of G12 with P[8] may be due to virus evolution through reassortment with an already established P type in this setting. This association of G12 strains with different P types can be considered part of its potential evolution toward becoming a stable strain.
The G12 strains described so far have been reported with various P types, such as P[4], P[6], P[8], P[9], or undetermined P types from different parts of the world. This is significant considering the evolution of G9 rotavirus strains. In India, a high incidence of G9P[11] rotavirus infections was initially described (4, 5). These strains were subsequently replaced by G9P[6] strains, which then probably reassorted with human strains of the P[8] VP4 genotype to become G9P[8], now a widely prevalent genotype in India (13). G9 strains have also now been identified as a major global genotype mainly as G9P[8] strains (14); however, these belong to a lineage different from the G9P[11] strains originally described in India.
It is known that nearly every child will experience at least one episode of rotavirus gastroenteritis by the age of 5 (20). Primary infections are believed to be more severe and to occur at a younger age in developing countries (18). In this study, severe rotavirus infection by a G12 strain was seen among children of >2 years of age. Four of the five children with symptomatic G12 infections in the community had previous rotavirus infections. Though the numbers described in this study are small, these infections raise the possibility that there may be no cross-protection against G12 strains from previous rotavirus infections. This may have significant implications for the efficacy of vaccines currently in use in different parts of the world, including recently licensed vaccines from Merck and Glaxo SmithKline and vaccines currently in development. The current vaccines do not include the G12 genotype, and it remains to be determined whether these vaccines will cross-protect against G12 infections. These data reinforce the necessity that a strain surveillance component be included in all countries for monitoring of circulating strains and detection of emerging rotaviruses before and after the introduction of a rotavirus vaccine.
Acknowledgments
This work was supported by the Wellcome Trust Trilateral Initiative for Infectious Diseases, grant no. 063144.
Footnotes
Published ahead of print on 29 November 2006.
REFERENCES
- 1.Banerjee, I., S. Ramani, B. Primrose, P. Moses, M. Iturriza-Gomara, J. J. Gray, S. Jaffar, B. Monica, J. P. Muliyil, D. W. Brown, and G. Kang. 2006. Comparative study of rotavirus epidemiology in children from a community-based birth cohort and a tertiary hospital in south India. J. Clin. Microbiol. 44:2468-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castello, A. A., M. H. Arguelles, R. P. Rota, A. Olthoff, B. Jiang, R. I. Glass, J. R. Gentsch, and G. Glikmann. 2006. Molecular epidemiology of group A rotavirus diarrhea among children in Buenos Aires, Argentina, from 1999 to 2003 and emergence of the infrequent genotype G12. J. Clin. Microbiol. 44:2046-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das, B., J. R. Gentsch, H. G. Cicirello, P. A. Woods, A. Gupta, M. Ramachandran, R. Kumar, M. K. Bhan, and R. I. Glass. 1994. Characterization of rotavirus strains from newborns in New Delhi, India. J. Clin. Microbiol. 32:1820-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das, B., J. R. Gentsch, Y. Hoshino, S. Ishida, O. Nakagomi, M. K. Bhan, R. Kumar, and R. I. Glass. 1993. Characterization of the G serotype and genogroup of New Delhi newborn rotavirus strain 116E. Virology 197:99-107. [DOI] [PubMed] [Google Scholar]
- 6.Das, S., V. Varghese, S. Chaudhury, P. Barman, S. Mahapatra, K. Kojima, S. K. Bhattacharya, T. Krishnan, R. K. Ratho, G. P. Chhotray, A. C. Phukan, N. Kobayashi, and T. N. Naik. 2003. Emergence of novel human group A rotavirus G12 strains in India. J. Clin. Microbiol. 41:2760-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estes, M. 1996. Rotaviruses and their replication, p. 1625-1655. In P. M. Howley, D. M. Knipe, and B. N. Fields (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 8.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z. Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffin, D. D., T. Nakagomi, Y. Hoshino, O. Nakagomi, C. D. Kirkwood, U. D. Parashar, R. I. Glass, and J. R. Gentsch. 2002. Characterization of nontypeable rotavirus strains from the United States: identification of a new rotavirus reassortant (P2A[6], G12) and rare P3[9] strains related to bovine rotaviruses. Virology 294:256-269. [DOI] [PubMed] [Google Scholar]
- 11.Iturriza-Gomara, M., J. Green, D. W. Brown, U. Desselberger, and J. J. Gray. 1999. Comparison of specific and random priming in the reverse transcriptase polymerase chain reaction for genotyping group A rotaviruses. J. Virol. Methods 78:93-103. [DOI] [PubMed] [Google Scholar]
- 12.Iturriza-Gomara, M., G. Kang, and J. Gray. 2004. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J. Clin. Virol. 31:259-265. [DOI] [PubMed] [Google Scholar]
- 13.Iturriza-Gomara, M., G. Kang, A. Mammen, A. K. Jana, M. Abraham, U. Desselberger, D. Brown, and J. Gray. 2004. Characterization of G10P[11] rotaviruses causing acute gastroenteritis in neonates and infants in Vellore, India. J. Clin. Microbiol. 42:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang, G. 2006. Rotavirus genotypes and severity of diarrheal disease. Clin. Infect. Dis. 43:315-316. [DOI] [PubMed] [Google Scholar]
- 15.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 16.Kulldorff, M. 1997. A spatial scan statistic. Commun. Stat. Theory Methods 26:1481-1496. [Google Scholar]
- 17.Kulldorff, M. R. K., G. Gherman, G. Williams, and D. DeFrancesco. 1998. Software for the spatial and space-time scan statistics: SaTScan. National Cancer Institute, Bethesda, MD.
- 18.Parashar, U. D., J. S. Bresee, J. R. Gentsch, and R. I. Glass. 1998. Rotavirus. Emerg. Infect. Dis. 4:561-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parashar, U. D., C. J. Gibson, J. S. Bresee, and R. I. Glass. 2006. Rotavirus and severe childhood diarrhea. Emerg. Infect. Dis. 12:304-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parashar, U. D., E. G. Hummelman, J. S. Bresee, M. A. Miller, and R. I. Glass. 2003. Global illness and deaths caused by rotavirus disease in children. Emerg. Infect. Dis. 9:565-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pongsuwanna, Y., R. Guntapong, M. Chiwakul, R. Tacharoenmuang, N. Onvimala, M. Wakuda, N. Kobayashi, and K. Taniguchi. 2002. Detection of a human rotavirus with G12 and P[9] specificity in Thailand. J. Clin. Microbiol. 40:1390-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruuska, T., and T. Vesikari. 1990. Rotavirus disease in Finnish children: use of numerical scores for clinical severity of diarrheal episodes. Scand. J. Infect. Dis. 22:259-267. [DOI] [PubMed] [Google Scholar]
- 23.Samajdar, S., V. Varghese, P. Barman, S. Ghosh, U. Mitra, P. Dutta, S. K. Bhattacharya, M. V. Narasimham, P. Panda, T. Krishnan, N. Kobayashi, and T. N. Naik. 2006. Changing pattern of human group A rotaviruses: emergence of G12 as an important pathogen among children in eastern India. J. Clin. Virol. 36:183-188. [DOI] [PubMed] [Google Scholar]
- 24.Santos, N., and Y. Hoshino. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev. Med. Virol. 15:29-56. [DOI] [PubMed] [Google Scholar]
- 25.Shinozaki, K., M. Okada, S. Nagashima, I. Kaiho, and K. Taniguchi. 2004. Characterization of human rotavirus strains with G12 and P[9] detected in Japan. J. Med. Virol. 73:612-616. [DOI] [PubMed] [Google Scholar]
- 26.Wakuda, M., S. Nagashima, N. Kobayashi, Y. Pongsuwanna, and K. Taniguchi. 2003. Serologic and genomic characterization of a G12 human rotavirus in Thailand. J. Clin. Microbiol. 41:5764-5769. [DOI] [PMC free article] [PubMed] [Google Scholar]