Abstract
Cytomegalovirus (CMV) is a significant cause of morbidity and mortality in lung transplant recipients (LTRs). The aim of the present study was to elucidate the relationship between the CMV DNA load in the lung compartment and that in plasma. For CMV load determination, the level of CMV DNA in plasma and bronchoalveolar lavage (BAL) samples was measured in a total of 97 paired BAL and plasma samples obtained from 25 LTRs. The original virus concentration in the epithelial lining fluid (ELF) was calculated from the BAL samples by correcting for dilution using the urea dilution method. In addition, the load of Epstein-Barr virus (EBV) and that of human herpesvirus 6 (HHV-6) DNA also were determined in BAL samples, recalculated for their concentrations in the ELF, and compared with the CMV DNA load. CMV DNA was found more frequently and at significantly higher levels in the lung compartment than in plasma (P < 0.001, Wilcoxon test), and the CMV load in the ELF was associated with symptomatic CMV disease. EBV and HHV-6 were detected in 43.6% and 21.7% of the ELF samples, respectively. A statistically significant association was found between the CMV and EBV DNA loads in the ELF (P < 0.001; Spearman's rho = 0.651). Thus, in LTRs, determination of the CMV DNA load in the lung compartment may be advantageous compared to monitoring only viremia. The significant relationship between EBV and CMV DNA loads in the ELF of LTRs and its clinical impact require further investigation.
Herpesvirus infections are a considerable burden and pose a challenge to the management of organ transplant recipients (16). Among herpesvirus infections, cytomegalovirus (CMV) infection is one of the most severe complications in organ transplant patients, and lung transplant recipients (LTRs) are at particularly high risk of developing CMV infection and disease (23). CMV detection in bronchoalveolar lavage (BAL) samples has been described as an earlier marker of virus replication in the lung than detection using PCR analysis of blood (4). However, CMV DNA quantitation from BAL samples has not been established as a routine predictive marker because a substantial overlap between virus loads in symptomatic and asymptomatic patients has been shown (4). This inability to discriminate symptomatic from asymptomatic LTRs by CMV DNA quantitation with BAL samples may be explained by the variability of the amount of epithelial lining fluid (ELF) which is recovered from the surface of the lung by BAL sampling. We have recently shown that in order to achieve an exact quantification of the CMV load in the lung compartment, corrections have to be made to account for the dilution of the original ELF containing the virus in the final BAL sample. Using this approach, we have found that the virus level in the lung compartment is clearly associated with CMV disease (27).
The number of studies in which interactions between herpesviruses in transplant patients have been investigated is limited, and data on viral coinfection in the lung compartment are especially rare. A few clinical studies have investigated whether there is an association between CMV and Epstein-Barr virus (EBV) reactivation in the blood compartment of immunosuppressed patients. While some authors (24, 21) have found that reactivation of each virus occurred independently, others (9, 11) have shown an association between CMV infection and the serologic profile of EBV reactivation. In vitro studies have shown as well that there might be an association between CMV and EBV (2).
Also, human herpesvirus 6 (HHV-6) has been increasingly recognized as a frequently detectable pathogen after transplantation (10, 15) and has been proposed as a possible cofactor for CMV disease leading to increased CMV replication in the blood compartment (17, 10, 5). Associations between EBV and HHV-6 also have been described, and concomitance of positive EBV and HHV-6 PCR results was shown with BAL samples (15).
The aim of the present study was to assess whether, for lung transplant recipients, there is a relationship between the CMV DNA load in the lung compartment, determined exactly by correcting for the BAL dilution factor, and the CMV DNA load in the blood. In addition, it was further investigated whether and at which concentrations EBV and HHV-6 DNA are present in the lung compartment and how they correlate with the CMV DNA level in ELF.
MATERIALS AND METHODS
Patients.
A total of 25 patients who had undergone lung transplantation at the Medical University hospital in Vienna were included in the study. The patient group consisted of 11 females and 14 males, with ages ranging from 23 to 68 years (median, 53.8 years) at the time of transplantation. Lung transplantation was performed because of chronic obstructive pulmonary disease (n = 16), primary pulmonary hypertension (n = 4), lung fibrosis (n = 3), sarcoidosis (n = 1), or lymphangioleiomyomatosis (n = 1).
The CMV serostatus was as follows: D+/R+ (D, donor; R, recipient), 11 patients; D−/R+, 7 patients; D+/R−, 2 patients. In five cases, the serostatus of either the donor, the recipient, or both was unknown. The LTRs were EBV seropositive in 12 cases, and in 13 cases the EBV serostatus was unknown.
During the time period investigated in this study, the immunosuppressive regimen administered to the patients consisted of steroids, cyclosporine or tacrolimus, and mycophenolate. As standard CMV prophylaxis, all patients received intravenous ganciclovir at a dosage of 10 mg/kg of body weight/day for 3 weeks after transplantation, followed by oral ganciclovir therapy at a dosage of 3 g/day until day 100 after transplantation. CMV disease was diagnosed according to criteria defined previously (26). Patients were categorized as asymptomatic in the study if they neither developed CMV disease nor received preemptive therapy for at least 3 months after sample collection.
BALs and plasma.
The BAL sampling procedure was carried out as described previously (27), and the corresponding plasma specimens were withdrawn simultaneously. A total of 97 simultaneously withdrawn BAL and plasma samples were analyzed retrospectively. All specimens were collected in the course of the routine follow-up in the years 1999 to 2003 and stored at −20°C. Thirty-two paired samples were obtained during the first 100 days after transplantation (TX) and 32 within 3 to 12 months after TX, and 33 were withdrawn later than 1 year after TX. The patients' clinical conditions at the time of sample withdrawal were as follows: 32 paired samples were obtained from LTRs receiving CMV prophylaxis, 29 were obtained from LTRs in an asymptomatic phase, 17 from LTRs receiving CMV therapy, and 13 from LTRs 1 to 5 days prior to the administration of ganciclovir therapy because of symptomatic CMV infection. The remaining six paired samples were obtained from asymptomatic LTRs within 3 months before initiation of anti-CMV therapy and were therefore not assigned to the asymptomatic group.
As controls, 41 BAL samples collected for routine diagnostic purposes from 41 immunocompetent patients with various lung diseases not associated with CMV disease were investigated for CMV, EBV, and HHV-6 DNA load. Patients with a history of transplantation, interstitial fibrosis, or human immunodeficiency virus infection were not included.
Methods. (i) Quantitative PCR. (a) CMV.
Sample extraction and quantitative PCR with BAL and plasma samples were performed using the semiautomated Cobas system (Roche Diagnostics, Branchburg, NJ) according to the manufacturer's instructions.
(b) EBV and HHV-6.
Sample extraction and EBV and HHV-6 quantitation with BAL and plasma samples by real-time PCR were performed as described previously using primers and a probe located within the EBNA1 gene and the HHV-6 U57 gene, respectively (1). The TaqMan PCR was carried out with a final volume of 50 μl using 10 μl template DNA extracted using a QIAamp viral RNA kit (QIAGEN, Hilden, Germany), 25 μl of TaqMan universal PCR master mix (PE Applied Biosystems, Foster City, CA), 25 pmol of each primer, and 10 pmol of each fluorescence-labeled probe with the fluorophore 6-carboxyfluorescein at the 5′ end and 6-carboxytetramethylrhodamine at the 3′ end. Amplification and detection were performed using the i-cycler iQ (Bio-Rad Laboratories, Hercules, CA) under the following conditions: 3 min at 50°C, 10 min at 95°C, and 45 cycles of 15 s at 95°C and either 30 s at 55°C/30 s at 72°C for EBV PCR or 1 min at 60°C for amplifying HHV-6 DNA. The detection limit of the EBV and HHV-6 PCR was found to be about 100 copies of viral DNA/ml.
(ii) Virus loads in the ELF.
To achieve an exact quantification of the CMV, EBV, and HHV-6 DNA loads in the lung compartment, the original virus concentration in the ELF, which is retrieved in diluted form by BAL sampling, was calculated using the urea dilution method (18, 27). In brief, the urea dilution method is based on the knowledge that urea is a solute whose concentrations in plasma and in the ELF are equal. The concentrations of urea in plasma and in the BAL sample taken in parallel were determined using the urease-glutamate dehydrogenase method using the Roche/Hitachi Modular Analytics system. Then, the virus load in the ELF was calculated using the urea dilution quotient.
(iii) Statistical analysis.
Statistical analysis was performed using the SPSS 11.5 software. The Wilcoxon matched-pairs test, the Mann-Whitney U test, and Spearman rank correlation analysis were performed where appropriate. A P value of <0.05 was regarded as statistically significant.
RESULTS
CMV DNA load in epithelial lining fluid and plasma.
In order to investigate the association between the CMV DNA load of the lung compartment and that of the plasma, 97 paired BAL and plasma samples were analyzed by CMV DNA PCR. Of the 97 BAL samples investigated, 51 (52.6%) were positive by PCR. The dilution of the original virus concentration in different BAL samples was found to vary considerably, with a range of between 2.47- and 320.72-fold. The calculated CMV load in the ELF ranged from 4.2 × 102 to 2.5 × 107 copies/ml. Of the corresponding plasma samples, 26 (26.8%) yielded positive results, showing viral loads between 1.4 × 101 to 7.0 × 104 copies/ml. Simultaneous detection of CMV in ELF and plasma occurred in 21 (21.6%) of the paired samples.
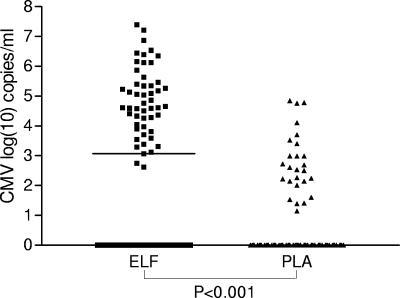
Comparison of the CMV DNA load in ELF and plasma was done for all 97 paired samples. As shown in Fig. 1, a significantly higher virus load was detected in the ELF than in plasma (P < 0.001, Wilcoxon matched pairs test). The ELF virus load was in each case but one found to be higher than the corresponding plasma virus load, with a difference of up to 6.87 logs.
FIG. 1.
Comparison of the CMV DNA levels in ELF and in simultaneously obtained plasma (PLA) samples (n = 97). Negative PCR results were set equal to 1 (log10 = 0). The bar indicates the median viral load (ELF, 1.2 × 10e3 copies/ml; plasma, negative).
CMV load in symptomatic versus asymptomatic patients.
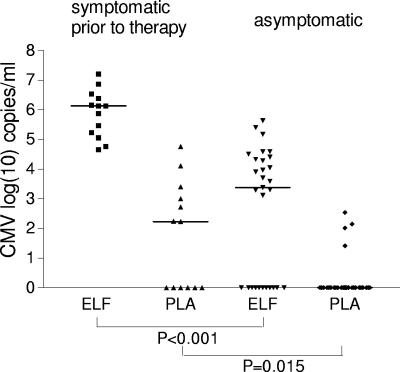
In order to compare the significance of the CMV DNA loads in ELF and in plasma as an early marker for CMV disease, the virus loads in both compartments were determined for symptomatic and asymptomatic LTRs. Thirteen paired samples obtained from patients at the onset of symptoms prior to therapy and 29 paired samples from patients in an asymptomatic phase were investigated. The results are presented in Fig. 2. The present data show that the calculated median virus load in the ELF was significantly higher in samples obtained from patients at the onset of symptomatic CMV disease prior to therapy than in those from asymptomatic patients (P < 0.001, Mann-Whitney U test).
FIG. 2.
Comparison of the CMV DNA load with the clinical status. Negative PCR results were set equal to 1 (log10 = 0). The bars indicate the median viral loads. Symptomatic patients prior to therapy (n = 13): ELF median, 1.3 × 10e6 copies/ml; PLA median, 1.7 × 10e2 copies/ml. Asymptomatic patients (n = 29): ELF median, 2.4 × 10e3 copies/ml; PLA median, negative.
Comparison of the plasma virus load in these two patient groups also revealed a significant mean difference (P = 0.015, Mann-Whitney U test) (Fig. 2). No CMV DNA was detectable in six of the plasma samples obtained from patients at the onset of symptomatic disease. The virus load in the corresponding ELFs ranged between 4.6 × 104 and 7.4 × 106 copies/ml, and all of these patients were clinically diagnosed as having CMV pneumonitis, according to a previous definition (12). Retesting the plasma samples using a different PCR method, an in-house real-time TaqMan PCR (1), confirmed these negative results.
EBV- and HHV-6 DNA load in the ELF.
To assess whether and at which concentrations other herpesviruses were present in the lung compartment of LTRs, quantitative EBV and HHV-6 DNA PCR assays were performed on the BAL samples in which sufficient material was still available for further analysis, and the viral DNA concentration in the ELF was calculated.
The EBV load was determined in 78 of the 97 BAL samples. Thirty-four (43.6%) of the BAL samples were positive by EBV DNA PCR, with a calculated viral load in the ELF ranging from 1.0 × 103 to 1.6 × 106 EBV DNA copies/ml (median, 3.2 × 104 copies/ml).
In 60 of the 78 BAL samples tested for CMV and EBV DNA load, the HHV-6 load was also measured. Thirteen (21.7%) of the BALs yielded a positive result, with the viral load in the ELF ranging from 1.5 × 102 to 3.2 × 104 copies/ml (median, 3.1 × 103 copies/ml).
Forty-one control BAL samples that were routinely withdrawn from patients in the course of various lung diseases (see Material and Methods) were also tested for the presence of herpesviruses. None of these samples was positive for CMV or HHV-6 DNA. EBV DNA was detected in 2 of the 41 samples (4.9%), with a viral load in the ELF of 3.8 × 103 and 2.7 × 104 EBV DNA copies/ml, respectively.
Association between CMV, EBV, and HHV-6 in the ELF.
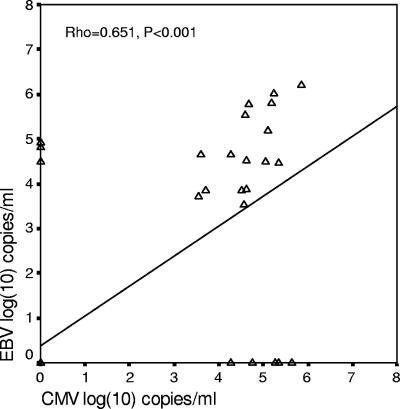
We then further investigated whether there is a relationship between the different herpesviruses in the lung compartment. Of the BAL samples, 33.3% were positive for both EBV and CMV DNA, 11.7% contained both CMV and HHV-6 DNA, and 11.7% were positive for both EBV and HHV-6 DNA. The quantitative data obtained for EBV and CMV, respectively, which were within the dynamic range of the assays (n = 62) were set in association (Fig. 3). Statistical analysis of the virus load levels in the individual samples showed a significant correlation between the EBV and CMV DNA concentrations in the ELF (Spearman's rho = 0.651; P < 0.001). For HHV-6, mostly low DNA levels were detected, and therefore no comparison with EBV and CMV loads was assessed. The simultaneous presence of CMV, EBV, and HHV-6 DNA was found in 6 of the 60 BAL samples (10.0%).
FIG. 3.
Association of CMV and EBV DNA loads in the ELF. Only samples with viral loads within the respective dynamic range of the quantitation assays are included (n = 62).
DISCUSSION
The data assessed in the present study show that the CMV DNA load in the lung compartment of lung transplant recipients is, in general, significantly higher than that in plasma, with a difference of up to 7 logs. A higher viral load was observed in the ELF independently of whether there was ongoing active CMV disease in the lung allograft (12) or not. It has been shown before that the lung is a major reservoir for CMV (3), and both alveolar epithelial cells and endothelial cells in the transplanted lung are fully permissive for CMV replication (14). The more frequent presence of CMV in the lung compartment than in plasma, as well as the remarkably higher ELF CMV DNA concentrations found in the patients, may thus be explained by a higher local level of viral replication in the lung. A higher virus production level in the lung has already been suggested by Riise et al. (19), who have reported high CMV DNA levels in the BAL fluid of lung transplant patients who did not show clinical symptoms of CMV disease, and this finding may possibly be a specific feature of CMV infection in LTRs.
We have previously shown that there is an association between a certain level of ELF CMV DNA load and symptomatic infection (27). This was confirmed again by the present data. In contrast to the previous findings, however, it was now also observed that in rare cases, asymptomatic patients also may harbor higher ELF virus loads.
The clinical importance of investigating the CMV DNA load in the lung compartment of LTRs was further underscored by the finding that CMV DNA was not detectable in 46.3% of the plasma samples obtained from patients at the onset of symptomatic CMV disease, while CMV DNA was present at high concentrations in all of the corresponding ELFs. Our results thus indicate that in LTRs, active CMV disease in the lung allograft may occur in the absence of detectable CMV DNAemia. This finding is in agreement with those of a previous study conducted by Chemaly et al. (7), who reported that in one out of five patients diagnosed with CMV pneumonitis, the plasma CMV PCR was negative while CMV DNA was present in all of the corresponding BAL samples. Similarly, Westall et al. (25) found that in two out of six patients with histological evidence of CMV infection of the lung, CMV DNA was absent in plasma while the BAL fluid was CMV PCR positive in all cases. Thus, routine CMV DNA load measurement in the lung compartment may provide an additional clinical advantage for lung transplant recipients.
The lower respiratory tract also has been suggested to be a major site of EBV latency (13), and in vivo EBV replication within lung alveolar cells has been demonstrated (8). The pathogenic role of EBV in pulmonary tissue, however, has not yet been sufficiently clarified (8). In the present study, EBV DNA was detected in 43.6% of the BAL samples obtained, and the recalculation for the ELF showed that high concentrations of EBV DNA of up to 1.6 × 106 copies/ml can be present in the ELF. Whether there is a direct clinical impact of the EBV DNA load in the lung compartment on the LTRs requires further analysis. EBV DNA was also found rarely (4.9%) in the control BALs. Since it has been reported before that EBV DNA can be detected in different tissues from healthy subjects, including the lungs (13), the occasional presence of EBV in BAL samples is not surprising. Our data show, however, that EBV DNA may be present in these nonimmunosuppressed patients at substantial viral loads in the ELF. The implications of this finding are also not yet clear.
Simultaneous infection with different herpesviruses has been observed especially for the blood compartment. When analyzing the BAL samples of our patients, a striking level of coexistence of EBV and CMV DNA was detected, occurring in one-third of the samples analyzed. In addition, a statistically significant association was found between the levels of CMV and the EBV DNA load in the ELF of the patients. These data indicate that there might be an association between CMV and EBV infection in the lung compartment. Different aspects could account for this association. First, the increase of EBV could be due to an increase in the number of cells in the BAL fluid and an expansion of latently EBV-infected B cells in the course of CMV infection of the lung allograft. Although Stephan et al. (20) reported a significant increase in the total lymphocyte population in BAL samples concurrent with pulmonary CMV infection, it was also found that B lymphocytes were almost entirely absent from BAL samples. Considering this, the simultaneous presence of EBV and CMV is not likely due to CMV-driven elevation of the total BAL cell count. A second explanation would be that there is an enhancement in virus replication due to the fact that both viruses replicate in the same cells. It has been shown that both EBV and CMV may infect alveolar epithelial cells, and productive replication of both herpesviruses may thus occur in the same cells (22, 14). Thus, EBV detected in the BAL could possibly be derived from lytic infection rather than from latently infected B cells.
HHV-6 DNA was detected in the ELF of the LTRs at clearly lower frequencies and concentrations than CMV or EBV and only in a limited part of the samples in coinfection with the other herpesviruses. HHV-6 has previously been shown to infect lung macrophages and epithelial cells (6), and its presence in BAL samples from LTRs has been demonstrated previously (15). But so far a possible pathogenic role of HHV-6 in LTRs has not been demonstrated.
In conclusion, our study shows that CMV DNA is generally more frequently present in the lung compartment than in plasma and at a clearly higher level in the ELF than in plasma. The CMV DNA load in the ELF is associated with symptomatic CMV disease, and determination of the CMV concentration in the lung compartment may be advantageous compared to monitoring of DNAemia in LTRs. It was also shown that in addition to CMV, EBV DNA is frequently present in the lung compartment of LTRs, and there is a significant association between the CMV and the EBV DNA loads in ELF. Further investigation will be required to see whether there is a clinical impact of herpesvirus coinfection in the lung compartment of lung transplant recipients.
Acknowledgments
We thank Barbara Dalmatiner, Claudia Kellner, and Katharina Leski for their excellent technical assistance.
Footnotes
Published ahead of print on 6 December 2006.
REFERENCES
- 1.Aberle, S. W., and E. Puchhammer-Stockl. 2002. Diagnosis of herpesvirus infections of the central nervous system. J. Clin. Virol. 25(Suppl. 1):S79-S85. [DOI] [PubMed] [Google Scholar]
- 2.Arcenas, R., and R. H. Widen. 2002. Epstein-Barr virus reactivation after superinfection of the BJAB-B1 and P3HR-1 cell lines with cytomegalovirus. BMC Microbiol. 2:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balthesen, M., M. Messerle, and M. J. Reddehase. 1993. Lungs are a major organ site of cytomegalovirus latency and recurrence. J. Virol. 67:5360-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buffone, G. J., A. Frost, T. Samo, G. J. Demmler, P. T. Cagle, and E. C. Lawrence. 1993. The diagnosis of CMV pneumonitis in lung and heart/lung transplant patients by PCR compared with traditional laboratory criteria. Transplantation 56:342-347. [DOI] [PubMed] [Google Scholar]
- 5.Cainelli, F., and S. Vento. 2002. Infections and solid organ transplant rejection: a cause-and-effect relationship? Lancet Infect. Dis. 2:539-549. [DOI] [PubMed] [Google Scholar]
- 6.Campadelli-Fiume, G., P. Mirandola, and L. Menotti. 1999. Human herpesvirus 6: an emerging pathogen. Emerg. Infect. Dis. 5:353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chemaly, R. F., B. Yen-Lieberman, J. Chapman, A. Reilly, B. N. Bekele, S. M. Gordon, G. W. Procop, N. Shrestha, C. M. Isada, M. Decamp, and R. K. Avery. 2005. Clinical utility of cytomegalovirus viral load in bronchoalveolar lavage in lung transplant recipients. Am. J. Transplant. 5:544-548. [DOI] [PubMed] [Google Scholar]
- 8.Egan, J. J., J. P. Stewart, P. S. Hasleton, J. R. Arrand, K. B. Carroll, and A. A. Woodcock. 1995. Epstein-Barr virus replication within pulmonary epithelial cells in cryptogenic fibrosing alveolitis. Thorax 50:1234-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hornef, M. W., G. Bein, L. Fricke, J. Steinhoff, H. J. Wagner, W. Hinderer, H. H. Sonneborn, and H. Kirchner. 1995. Coincidence of Epstein-Barr virus reactivation, cytomegalovirus infection, and rejection episodes in renal transplant recipients. Transplantation 60:474-480. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs, F., C. Knoop, F. Brancart, P. Gilot, C. Melot, B. Byl, M. L. Delforge, M. Estenne, and C. Liesnard. 2003. Human herpesvirus-6 infection after lung and heart-lung transplantation: a prospective longitudinal study. Transplantation 75:1996-2001. [DOI] [PubMed] [Google Scholar]
- 11.Khameneh, Z. R., J. Soin, M. Durlik, M. Lao, L. Paczek, and Z. Gaciong. 1999. Factors affecting reactivation of Epstein-Barr virus infection after kidney allograft transplantation. Ann. Transplant. 4:18-22. [PubMed] [Google Scholar]
- 12.Ljungman, P., and S. A. Plotkin. 1995. Workshop on CMV disease: definitions, clinical severity scores, and new syndromes. Scand. J. Infect. Dis. Suppl. 99:87-89. [Google Scholar]
- 13.Lung, M. L., W. K. Lam, S. Y. So, W. P. Lam, K. H. Chan, and M. H. Ng. 1985. Evidence that respiratory tract is major reservoir for Epstein-Barr virus. Lancet i:889-892. [DOI] [PubMed] [Google Scholar]
- 14.Morbini, P., and E. Arbustini. 2001. In situ characterization of human cytomegalovirus infection of bronchiolar cells in human transplanted lung. Virchows Arch. 438:558-566. [DOI] [PubMed] [Google Scholar]
- 15.Neurohr, C., P. Huppmann, H. Leuchte, M. Schwaiblmair, I. Bittmann, G. Jaeger, R. Hatz, L. Frey, P. Uberfuhr, B. Reichart, and J. Behr. 2005. Human herpesvirus 6 in bronchalveolar lavage fluid after lung transplantation: a risk factor for bronchiolitis obliterans syndrome? Am. J. Transplant. 5:2982-2991. [DOI] [PubMed] [Google Scholar]
- 16.Razonable, R. R., and C. V. Paya. 2003. Herpesvirus infections in transplant recipients: current challenges in the clinical management of cytomegalovirus and Epstein-Barr virus infections. Herpes 10:60-65. [PubMed] [Google Scholar]
- 17.Razonable, R. R., R. A. Brown, A. Humar, E. Covington, E. Alecock, and C. V. Paya. 2005. Herpesvirus infections in solid organ transplant patients at high risk of primary cytomegalovirus disease. J. Infect. Dis. 192:1331-1339. [DOI] [PubMed] [Google Scholar]
- 18.Rennard, S. I., G. Basset, D. Lecossier, K. M. O'Donnell, P. Pinkston, P. G. Martin, and R. G. Crystal. 1986. Estimation of volume of epithelial lining fluid recovered by lavage using urea as marker of dilution. J. Appl. Physiol. 60:532-538. [DOI] [PubMed] [Google Scholar]
- 19.Riise, G. C., R. Andersson, T. Bergstrom, A. Lundmark, F. N. Nilsson, and S. Olofsson. 2000. Quantification of cytomegalovirus DNA in BAL fluid: a longitudinal study in lung transplant recipients. Chest 118:1653-1660. [DOI] [PubMed] [Google Scholar]
- 20.Stephan, F., J. F. Bernaudin, D. Cesari, A. Fajac, D. D. Grenet, I. Caubarrere, and M. Stern. 2001. Blood and alveolar lymphocyte subsets in pulmonary cytomegalovirus infection after lung transplantation. BMC Infect. Dis. 1:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevens, S. J., E. A. Verschuuren, I. Pronk, W. van Der Bij, M. C. Harmsen, T. H. The, C. J. Meijer, A. J. van Den Brule, and J. M. Middeldorp. 2001. Frequent monitoring of Epstein-Barr virus DNA load in unfractionated whole blood is essential for early detection of posttransplant lymphoproliferative disease in high-risk patients. Blood 97:1165-1171. [DOI] [PubMed] [Google Scholar]
- 22.Stewart, J. P., J. J. Egan, A. J. Ross, B. G. Kelly, S. S. Lok, P. S. Hasleton, and A. A. Woodcock. 1999. The detection of Epstein-Barr virus DNA in lung tissue from patients with idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 159:1336-1341. [DOI] [PubMed] [Google Scholar]
- 23.van der Bij, W., and R. Speich. 2001. Management of cytomegalovirus infection and disease after solid-organ transplantation. Clin. Infect. Dis. 33(Suppl. 1):S32-S37. [DOI] [PubMed] [Google Scholar]
- 24.Wagner, H. J., L. Fischer, W. J. Jabs, M. Holbe, K. Pethig, and P. Bucsky. 2002. Longitudinal analysis of Epstein-Barr viral load in plasma and peripheral blood mononuclear cells of transplanted patients by real-time polymerase chain reaction. Transplantation 74:656-664. [DOI] [PubMed] [Google Scholar]
- 25.Westall, G. P., A. Michaelides, T. J. Williams, G. I. Snell, and T. C. Kotsimbos. 2004. Human cytomegalovirus load in plasma and bronchoalveolar lavage fluid: a longitudinal study of lung transplant recipients. J. Infect. Dis. 190:1076-1083. [DOI] [PubMed] [Google Scholar]
- 26.Winston, D. J., and R. W. Busuttil. 2003. Randomized controlled trial of oral ganciclovir versus oral acyclovir after induction with intravenous ganciclovir for long-term prophylaxis of cytomegalovirus disease in cytomegalovirus-seropositive liver transplant recipients. Transplantation 75:229-233. [DOI] [PubMed] [Google Scholar]
- 27.Zedtwitz-Liebenstein, K., P. Jaksch, C. Bauer, T. Popow, W. Klepetko, H. Hofmann, and E. Puchhammer-Stockl. 2004. Association of cytomegalovirus DNA concentration in epithelial lining fluid and symptomatic cytomegalovirus infection in lung transplant recipients. Transplantation 77:1897-1899. [DOI] [PubMed] [Google Scholar]