Abstract
Globicatella sanguinis is a very rare isolate in clinical samples. We present a case of meningitis in a 69-year-old female patient after implantation of an external left ventricular drainage due to a hydrocephalus. She recovered after antibiotic treatment with ceftriaxone.
CASE REPORT
In October 2005 a 69-year-old female patient presented with a 1-year history of worsening weakness of her left leg and left arm and increasing problems with walking. She complained of being confused sometimes. Additionally, she had a longstanding history of depressive disorders for which she had been in psychiatric care for several years.
A computed tomography brain scan showed a dilated ventricular system, and the preliminary diagnosis of normal-pressure hydrocephalus was made. To confirm the diagnosis and to assess the suitability of the implantation of a ventriculoperitoneal shunt, an external left ventricular drainage was implanted on 26 October 2005. The diagnosis could be confirmed, and the drainage was removed on 28 October 2005. In the morning of 30 October 2005 the patient was somnolent, threw up several times, and presented clinical signs of meningitis. A control computed tomography scan showed an unchanged ventricular configuration. The differential diagnosis of decompensated hydrocephalus or infectious meningitis was made, and a new external ventricular drainage was implanted. In a sample of cerebrospinal fluid, lactate levels were elevated to 6.35 mmol/liter, and subsequently a gram-positive coccus was grown. The patient was put on intravenous ceftriaxone (2 g once daily for 10 days) and improved rapidly. After negative cerebrospinal fluid cultures, a definitive ventriculoperitoneal shunt was implanted on 9 November 2005. On the 22 November the patient was transferred to a specialized rehabilitation facility.
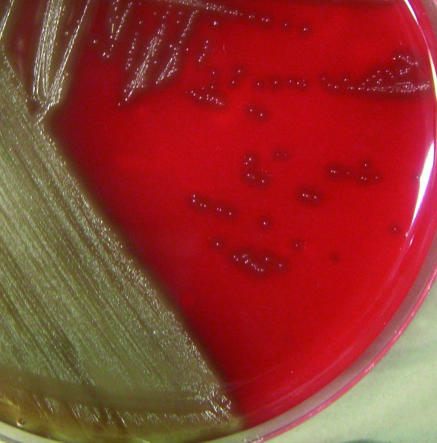
The culture of the first cerebrospinal fluid sample (Columbia agar supplemented with 5% sheep blood and pyridoxal, 37°C, 5%CO2) resulted in faint growth of rough, alpha-hemolytic colonies after 24 h, which was more pronounced after 48 h (Fig. 1). Gram staining showed a gram-positive coccus (short chains). A catalase test was negative. An optochin test (5-μg disk; Oxoid) was negative (no inhibition zone). The rapid ID32strep test resulted in the finding of a Globicatella sp. The Phoenix system (PMIC/ID-56) confirmed the API result and identified the bacterium as Globicatella sanguinis (certainty of 99%). Surprisingly, a 16S rRNA sequence analysis showed 99% identity with the 16S rRNA sequence of G. sulfidifaciens (GenBank accession no. AJ297627) and 95% identity with the16S rRNA sequence of G. sanguinis (GenBank accession no. S50214).
FIG. 1.
Globicatella sanguinis (48 h of growth, CO2, 37°C, Columbia agar supplemented with 5% sheep blood and pyridoxal).
Looking into the literature, we found that the biochemical profile of G. sanguinis is based on a total of 29 isolates (1a, 3). The biochemical profile of G. sulfidifaciens is based on a total of eight isolates (7). Of 33 reactions for our isolate, only one was not in line with G. sanguinis, but six reactions were not in line with G. sulfidifaciens, one of which is the production of sulfide (Table 1). In the original publication (7), the authors stressed the importance of this parameter for the differentiation between G. sanguinis (negative) and G. sulfidifaciens (positive). Our isolate is negative. On the other hand, there is only one 16S rRNA sequence for each Globicatella sp. in GenBank. Additionally, these sequences are very similar, and there is evidence that currently the database for the identification of Globicatella spp. based on 16S rRNA is too small to be helpful (2). Therefore, we named our isolate G. sanguinis in accordance with the biochemical profile.
TABLE 1.
Differences in biochemical profiles of G. sulfidifaciens, G. sanguinis, and our straina
| Test | Resulta for:
|
||
|---|---|---|---|
| G. sulfidifaciens | G. sanguinis | Our strain | |
| Mannitol | Neg | Pos | Weakly pos |
| β-Galactosidase | Neg | Pos | Pos |
| β-Glucuronidase | Pos | Neg | Neg |
| Hippurat | Neg | Pos | Pos |
| H2S (Kligler) | Pos | Neg | Neg |
| Arginine dihydrolase | ND | ND | Neg |
| β-Glucosidase | ND | ND | Neg |
| Sorbitol | Neg | Pos | Weakly pos |
| Lactose | Neg | Pos | Neg |
| α-Galactosidase | Pos | ND | Pos |
| Alkaline phosphatase | Neg | ND | Neg |
| Ribose | ND | Pos | Pos |
| Trehalose | Pos | Pos | Pos |
| Raffinose | Pos | Pos | Pos |
| Saccharose | Pos | ND | Pos |
| l-Arabinose | Var | ND | Pos |
| d-Arabitol | Neg | ND | Neg |
| Cyclodextrin | ND | ND | Neg |
| Voges-Proskauer | Neg | ND | Neg |
| Alanine-phenylalanine-proline arylamidase | ND | ND | Pos |
| β-Galactosidase | Var | ND | Pos |
| Pyroglutamic acid arylamidase | ND | ND | Pos |
| N-Acetyl-β-glucosamidase | ND | ND | Pos |
| Glycyl-tryptophane arylamidase | ND | ND | Neg |
| Glycogen | Pos | ND | Pos |
| Pullulan | ND | ND | Pos |
| Maltose | Pos | Pos | Pos |
| Melibiose | Pos | Pos | Pos |
| Melezitose | Neg | ND | Neg |
| Methyl-β-d-glucopyranoside | ND | ND | Neg |
| Tagatose | Neg | ND | Neg |
| β-Mannosidase | ND | ND | Neg |
| Urease | Neg | ND | Neg |
ND, not done; Pos, positive; Neg; negative; Var, variable. Results for our strain were determined by API 32strep and Phoenix testing. Data for G. sulfidifaciens and G. sanguinis are from references 7 and 1a, respectively. Boldface indicates differences between results for our strain and for G. sulfidifaciens.
Antimicrobial susceptibility testing was done according to the 2005 CLSI guidelines (1), using the established breakpoints for Streptococcus spp. other than S. pneumoniae. Broth microdilution was performed for penicillin G, cefotaxime, clarithromycin, clindamycin, tetracycline, telithromycin, gatifloxacin, and levofloxacin. MICs of vancomycin, linezolid, imipenem, erythromycin, and ciprofloxacin were determined using the Etest. The results are shown in Table 2.
TABLE 2.
MICs
| Antimicrobial agent | MIC (μg/ml) | Interpretationa |
|---|---|---|
| Penicillin G | 0.06 | Sensitive |
| Cefotaxime | 0.5 | Sensitive |
| Imipenem | 0.25 | No breakpoint |
| Erythromycin | 2 | Resistant |
| Clarithromycin | 1 | Resistant |
| Telithromycin | ≤0.03 | No breakpoint |
| Clindamycin | ≤0.12 | Sensitive |
| Ciprofloxacin | 0.38 | No breakpoint |
| Levofloxacin | <0.25 | Sensitive |
| Gatifloxacin | ≤0.06 | No breakpoint |
| Vancomycin | 0.125 | Sensitive |
| Tetracycline | 1.0 | Sensitive |
| Linezolid | 0.5 | Sensitive |
According to reference 1.
There are two well-known mechanisms for macrolide resistance. The presence of the mef gene is associated with macrolide-only resistance, whereas the ermB gene causes combined macrolide/lincosamide resistance. There are no published reports on the origin of macrolide resistance in Globicatella isolates. To investigate the resistance mechanism in our isolate, a mef PCR and an ermB PCR were performed (4, 5). The mef PCR was positive, whereas the ermB PCR was negative. These results confirm the macrolide resistance and the lincosamide sensitivity of our isolate.
G. sanguinis was first described in 1992 by Collins and coworkers, who named it G. sanguis (1a). It was renamed G. sanguinis in 1997 by Trüper and de'Clari (6). A PubMed search for Globicatella revealed that G. sanguinis was described as a cause of meningoencephalitis in lambs by Vela and coworkers in 2000 (8). Whether our patient had contact with sheep could not be elucidated. In 2001, Shewmaker and coworkers (3) published the first susceptibility testing results for G. sanguinis, which are in good accordance with our own results. Interestingly, 48% of their 27 strains showed resistance to cefotaxime, with a MIC50 of 1.0 mg/liter and a MIC90 of 4.0 mg/liter. Fortunately for our patient, our isolate was susceptible to cefotaxime.
Acknowledgments
We thank B. Weidenhaupt for excellent laboratory work.
Footnotes
Published ahead of print on 22 November 2006.
REFERENCES
- 1.CLSI. 2005. Performance standards for antimicrobial susceptibility testing, 15th informational supplement. M100-S15. CLSI, Wayne, Pa.
- 1a.Collins, M. D., M. Aguirre, R. R. Facklam, J. Shallcross, and A. M. Williams. 1992. Globicatella sanguis gen.nov., sp.nov., a new gram-positive catalase-negative bacterium from human sources. J. Appl. Bacteriol. 73:433-437. [DOI] [PubMed] [Google Scholar]
- 2.Lau, S. K., P. C. Woo, N. K. Li, J. L. Teng, K. W. Leung, K. H. Ng, T. L. Que, and K. Y. Yuen. 2006. Globicatella bacteraemia identified by 16S ribosomal RNA gene sequencing. J. Clin. Pathol. 59:303-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shewmaker, P. L., A. G. Steigerwalt, L. Shealey, R. Weyant, and R. R. Facklam. 2001. DNA relatedness, phenotypic characteristics, and antimicrobial susceptibilities of Globicatella sanguinis strains. J. Clin. Microbiol. 39:4052-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trieu-Cuot, P., C. Poyart-Salmeron, C. Carlier, and P. Courvalin. 1990. Nucleotide sequence of the erythromycin resistance gene of the conjugative transposon Tn1545. Nucleic Acids Res. 18:3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trüper, H. G., and L. de'Clari. 1997. Taxonomic note: necessary correction of specific epithets formed as substantives (nouns) “in apposition.” Int. J. Syst. Bacteriol. 47:908-909. [Google Scholar]
- 7.Vandamme, P., J. Hommez, C. Snauwaert, B. Hoste, I. Cleenwerck, K. Lefebvre, M. Vancanneyt, J. Swings, L. A. Devriese, and F. Haesebrouck. 2001. Globicatella sulfidifaciens sp. nov., isolated from purulent infections in domestic animals. Int. J. Syst. Evol. Microbiol. 51:1745-1749. [DOI] [PubMed] [Google Scholar]
- 8.Vela, A. I., E. Fernandez, A. las Heras, P. A. Lawson, L. Dominguez, M. D. Collins, and J. F. Fernandez-Garayzabal. 2000. Meningoencephalitis associated with Globicatella sanguinis infection in lambs. J. Clin. Microbiol. 38:4254-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]