Abstract
Clinical strains of Mycobacterium tuberculosis can be divided into three principal genetic groups based on the single-nucleotide polymorphisms at the katG gene codon 463 and the gyrA gene codon 95. One subgroup of genetic group 1, the Beijing/W lineage, has been widely studied because of its worldwide distribution and association with outbreaks. In order to increase our understanding of the clinical and epidemiological relevance of the genetic grouping of M. tuberculosis clinical strains and the Beijing/W lineage, we investigated the genetic grouping of 679 clinical isolates of M. tuberculosis, representing 96.3% of culture-confirmed tuberculosis cases diagnosed in Arkansas between January 1996 and December 2000 using PCR and DNA sequencing. We assessed the associations of infections by different genetic groups of M. tuberculosis strains and infection by the Beijing/W lineage strains with the clinical and epidemiological characteristics of the patients using chi-square tests and multivariate logistic regression analysis. Of the 679 study isolates, 676 fell into one of the three principal genetic groups, with 63 (9.3%) in group 1, 438 (64.8%) in group 2, and 175 (25.9%) in group 3. After adjusting for potential confounding of age, gender, race/ethnicity, human immunodeficiency virus serostatus, and plcD genotype in a multivariate logistic regression model, patients infected by the Beijing/W lineage isolates were nearly three times as likely as patients infected with the non-Beijing/W lineage isolates to have an extrathoracic involvement (odds ratio [95% confidence interval], 2.85 [1.33, 6.12]). Thus, the Beijing/W lineage strains may have some special biological features that facilitate the development of extrathoracic tuberculosis.
The integration of molecular analysis of Mycobacterium tuberculosis with an epidemiological analysis of clinical information provides a new tool to assess possible associations between M. tuberculosis strain type and the clinical and epidemiological characteristics of the disease (7, 15, 16, 28). Clinical strains of M. tuberculosis complex, the causative pathogen of tuberculosis (TB), can be divided into three principal genetic groups based on the combination of single-nucleotide polymorphisms (SNPs) that occur at codon 463 of the katG gene (katG463) encoding catalase-peroxidase and codon 95 of the gyrA gene (gyrA95) encoding the subunit A of DNA gyrase (22). However, our understanding of the clinical and epidemiological relevance of such genetic grouping is limited. Three previous studies conducted in two different regions of the United States and one region in Italy explored the association between genetic grouping and DNA fingerprint clustering of M. tuberculosis (8, 21, 22) that has been commonly used as a measurement for TB transmission (1). However, the findings of these studies were inconsistent. Two studies showed that the clustered cases were more likely to be caused by isolates in groups 1 and 2 than those in group 3, while the other study failed to demonstrate an association. These studies have used different definitions for clusters. Furthermore, not all the studies had access to a comprehensive patient database that would allow a thorough assessment of the relationship between the genetic grouping of the study isolates and the clinical and epidemiological characteristics of the study patients (8, 21, 22).
The Beijing/W lineage of M. tuberculosis is a subgroup of the principal genetic group 1 of particular clinical and epidemiologic interest (3, 13, 22). Isolates are defined as Beijing/W lineage strains based on their distinct spoligotyping pattern, i.e., the presence of at least three of the spacers 35 to 43 used for spoligotyping and the absence of spacers 1 to 34 (17). The Beijing/W lineage strains are dominant in Asia (26), and their spread has been documented worldwide (12). The Beijing/W lineage strains were found to be related to outbreaks (3). Patients infected with the Beijing genotype strains were found to be more likely to have a febrile response than patients infected with non-Beijing genotype strains shortly after the start of treatment (25). In addition, the Beijing genotype strains have been found to elicit a nonprotective immune response in mice (19). These findings suggest that the Beijing/W lineage strains may have specific pathogenic features. By contrast, a study using a selected sample in The Netherlands did not find significant differences in chest X-ray presentations between patients infected with the Beijing genotype strains and patients infected with other strains (4). Thus, the clinical relevance of the Beijing/W lineage strain infection remains to be clarified.
Current TB control strategy and transmission models are based on the assumption that all M. tuberculosis clinical strains are equally transmitted and virulent (22). However, if strains belonging to different genetic groups or lineages have different biological attributes producing different epidemiological and clinical phenotypes, public health control strategies might be tailored accordingly. Thus, a better understanding of the clinical and epidemiological relevance of M. tuberculosis genetic grouping in general and the Beijing/W lineage in particular may allow the development of better strategies for TB control. In this study, we explored the epidemiologic and clinical relevance of the principal genetic groups and the Beijing/W lineage of M. tuberculosis, using an integrated approach that combines comparative genomics and epidemiological data analysis and a 5-year population-based, epidemiologically and clinically well-characterized isolate collection.
MATERIALS AND METHODS
Study sample and patient data.
One M. tuberculosis isolate from each of 679 culture-confirmed TB patients diagnosed in Arkansas between 1 January 1996 and 31 December 2000 was used in this study. The study sample included all available isolates, representing 96.3% of all the culture-confirmed cases (705) diagnosed during the study period. Patient demographics and social behaviors as well as clinical characteristics of the diseases were obtained from the surveillance records of the Arkansas Department of Health. No statistically significant difference in the distribution of the variables used was found between the study sample and the excluded 26 cases. Genomic DNA was extracted from Lowenstein-Jensen slant cultures using standard procedures (20). The isolates were defined as clustered versus unique by a combination of the IS6110 restriction fragment-length polymorphism (RFLP) analysis and the pTBN12 secondary fingerprinting using the definition described previously (2, 6). Spoligotyping results were available for 675 isolates. Thirty-seven of these isolates were classified as the Beijing/W lineage strains based on the Beijing/W lineage-specific spoligotype S00034. The genotypes of the four plc genes of all isolates determined in a previous study were also available to this study (16, 28). The study protocols for human subject protection were approved by the Health Sciences Institutional Review Boards of the University of Michigan and the University of Arkansas for Medical Sciences.
PCR and DNA sequencing.
The SNPs at katG463 and gyrA95 were investigated by PCR assay and sequencing described previously (24). Double-stranded sequences were obtained for each PCR product using the PCR primers. Sequence comparison was performed using EditSeq 5.02 and MegAlign 5.01 software (DNAStar Inc., Madison, WI). Isolates were assigned to one of the three principal genetic groups based on the SNPs found in katG463 and gyrA95 as described previously (22).
Statistical analysis.
Two different analytical comparisons for answering two related but different questions were performed. We first analyzed the distribution of patient demographics among the three principal genetic groups to assess whether or not the infection of M. tuberculosis isolates of a given genetic group or a specific genetic lineage is associated with a given demographic or clinical/epidemiological category of the patients using the chi-square (χ2) test or Fisher's exact test, as appropriate. A stratified χ2 test was used to adjust for confounding factors, as there were only two variables that were considered to be confounders. Second, we analyzed the association between genetic grouping of the infecting isolates and the clinical characteristics using a multivariate logistic regression model to adjust for potential confounders. Potentially confounding variables included age, which was found to be associated with genetic grouping in the first analytic comparison, and several previously identified risk factors for extrathoracic TB, including human immunodeficiency virus (HIV) seropositive status, non-Hispanic black race/ethnicity, being female, and the infection of M. tuberculosis isolates with a mutant plcD gene (16, 28).
The disease sites were classified into thoracic and extrathoracic using the definitions described previously (28). Briefly, thoracic TB was defined as disease sites confined to the lung, pleura, and intrathoracic lymph nodes, while the extrathoracic TB were cases having extrathoracic disease with or without concurrent disease within the thoracic cavity. Considering the likelihood of relatedness among the cases within the same cluster defined by IS6110 RFLP and pTBN12 pattern, we used generalized estimating equations (GEE) (18, 29) to control for potential intracluster dependence when we assessed the associations between the genetic grouping of M. tuberculosis isolates and the clinical characteristics of the disease and the associations between the Beijing/W lineage infection and the clinical characteristics of the disease. GEE takes the within-cluster similarity into account when calculating standard errors of the regression parameters. The magnitude of the associations was estimated using the odds ratio (OR) and 95% confidence intervals (95% CI). Logistic regression models with GEE were used to control for potential confounders. All the statistical analyses were done using SAS version 8.0 (SAS Institute, Cary, NC).
RESULTS
Distribution of three principal genetic groups.
Six hundred seventy-six of the 679 study isolates were classifiable into one of the three principal genetic groups. Sequencing of these PCR products showed the presence of the three previously reported combinations of katG463 and gyrA95 polymorphisms. Among the 676 isolates, 63 (9.3%), 438 (64.8%), and 175 (25.9%) were classified into principal groups 1, 2, and 3, respectively. The remaining three isolates failed to generate a PCR product containing katG463, and one of the three isolates also failed to generate a product in PCR targeting gyrA95. The suitability of the DNA of this isolate for PCR was affirmed by amplification of the 16S rRNA gene.
Comparison of genetic diversity among three principal genetic groups.
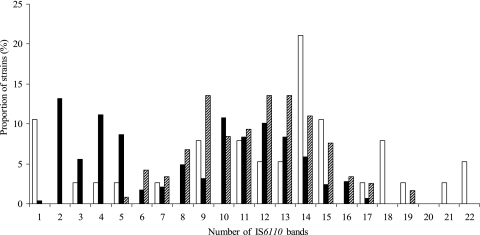
The frequency distribution of IS6110 copy number determined using 353 unique isolates and 91 isolates randomly selected from each of the 91 clusters differed among the three genetic groups (Fig. 1). Isolates in group 1 had the broadest range of IS6110 copy numbers. There were no isolates having less than five copies of IS6110 in group 3 and no isolates having more than 17 copies of IS6110 in group 2. Single-copy isolates were found almost exclusively in group 1. Group 1 had more genetic variations in the plcA, plcB, plcC, and plcD genes than those in group 2 or 3 (Table 1).
FIG. 1.
Distribution of IS6110 band numbers among the three principal genetic groups based on the analysis of 444 strains obtained from Arkansas between 1 January 1996 and 31 December 2000. x axis, number of the IS6110 hybridizing bands; y axis, proportion of strains in each of the three principal genetic groups by IS6110 copy number. The white bar represents group 1; the black bar represents group 2; and the hatched bar represents group 3.
TABLE 1.
Frequency distribution of low-copy (≤6) and high-copy (>6) IS6110 RFLPs and polymorphisms of the plc genes among the three principal genetic groups of M. tuberculosis strains obtained from Arkansas between 1 January 1996 and 31 December 2000 (n = 444)
| Characteristic of strains | No. (%) in group:
|
P valuea | ||
|---|---|---|---|---|
| 1 (n = 38) | 2 (n = 288) | 3 (n = 118) | ||
| No. of IS6110 copies | <0.0001 | |||
| ≤6 | 7 (18.4) | 117 (40.6) | 6 (5.1) | |
| >6 | 31 (81.6) | 171 (59.4) | 112 (94.9) | |
| plcD genotype | <0.0001 | |||
| plcD wild type | 19 (50.0) | 198 (68.8) | 75 (63.6) | |
| plcD mutant | 1 (2.6) | 10 (3.5) | 19 (16.1) | |
| plcD and adjacent genes mutant | 18 (47.4) | 80 (27.8) | 24 (20.3) | |
| plcC genotype | 0.0013 | |||
| plcC wild type | 31 (81.6) | 277 (96.2) | 110 (93.2) | |
| plcC mutant | 7 (18.4) | 11 (3.8) | 8 (6.8) | |
| plcB genotypes | <0.0001 | |||
| plcB wild type | 26 (68.4) | 282 (97.9) | 115 (97.5) | |
| plcB mutant | 12 (31.6) | 6 (2.1) | 3 (2.5) | |
| plcA genotypes | 0.0006 | |||
| plcA wild type | 31 (81.6) | 276 (95.8) | 114 (96.6) | |
| plcA mutant | 7 (18.4) | 12 (4.2) | 4 (3.4) | |
P values are from bivariate χ2 test or Fisher's exact test.
Distribution of selected patient demographics among the three genetic groups.
We observed statistically significant differences in the distributions of the patients' race/ethnicity and age among the three principal genetic groups (Table 2). Compared with patients infected by groups 2 and 3, patients infected by the group 1 isolates were more likely to be of Asian origin (for group 1 versus groups 2: OR [95% CI], 64.94 [23.58, 178.81]; for group 1 versus group 3: OR [95% CI], 64.87 [14.76, 285.09]) and were less likely to be 65 or older (for group 1 versus group 2: OR [95% CI], 0.22 [0.11, 0.41]; for group 1 versus group 3: OR [95% CI], 0.18 [0.09, 0.37]).
TABLE 2.
Frequency distribution of patient age and race/ethnicity among the three principal genetic groups of clinical isolates of M. tuberculosis (n = 676) obtained from Arkansas between 1 January 1996 and 31 December 2000
| Characteristic of patients | No. (%) in group:
|
P valuea | ||
|---|---|---|---|---|
| 1 (n = 63) | 2 (n = 438) | 3 (n = 175) | ||
| Age (yr) | <0.0001 | |||
| 0-14 | 0 (0.0) | 6 (1.4) | 0 (0.0) | |
| 15-24 | 6 (9.5) | 13 (3.0) | 2 (1.1) | |
| 25-44 | 20 (31.8) | 91 (20.8) | 34 (19.4) | |
| 45-64 | 25 (39.7) | 99 (22.6) | 41 (23.4) | |
| 65+ | 12 (19.0) | 229 (52.3) | 98 (56.0) | |
| Race/ethnicity | <0.0001 | |||
| Non-Hispanic white | 26 (41.3) | 252 (57.5) | 101 (57.7) | |
| Non-Hispanic black | 9 (14.3) | 159 (36.3) | 65 (37.1) | |
| Hispanic | 1 (1.6) | 19 (4.3) | 7 (4.0) | |
| Asian/Pacific Islander | 27 (42.9) | 5 (1.1) | 2 (1.1) | |
| American Indian | 0 (0.0) | 3 (0.7) | 0 (0.0) | |
P values are from bivariate χ2 test or Fisher's exact test.
Further analyses of the age distribution among the three genetic groups were restricted to non-Asian/Pacific Islanders to examine if the age distribution among the three genetic groups was confounded by Asian origin. Significant differences in the age distribution among the three principal genetic groups were also found in non-Asian/Pacific Islanders (P = 0.0095). The proportion of people older than 65 in non-Asian/Pacific Islanders was 25.0% in group 1, 52.9% in group 2, and 56.7% in group 3. In addition, the race/ethnicity distribution among the three genetic groups was analyzed, stratifying by age, to examine if the different race/ethnicity distribution among the three genetic groups described above had been confounded by age. Significant differences in the race/ethnicity distribution among the three principal genetic groups were found in both patients aged 65 or older (P < 0.0001) and in patients younger than 65 (P < 0.0001). Among people younger than 65, the proportions of Asian origin in genetic groups 1, 2, and 3 were 47.1%, 2.4%, and 2.6%, respectively; among those aged 65 or older, the proportions of Asian origin were 25.0%, 0.0%, and 0.0% in groups 1, 2, and 3, respectively.
Distribution of clinical characteristics.
Of the 676 isolates classifiable into one of the three principal genetic groups, 78 were related to cases with an extrathoracic involvement and 598 were related to thoracic cases. Of the 630 patients who had thoracic involvement, 579 had chest radiographs available for the study and 581 had sputum smear results. Based on the chest radiographs, 215 cases had cavitation in the lungs. Of the 581 cases with sputum smear results, 250 were smear positive. The distribution of three clinical characteristics, including site of disease, cavitation in the lungs, and sputum smear positivity, among the three genetic groups were analyzed using a χ2 test or Fisher's exact test. No significantly different distribution of cavitation in the lungs or positive sputum smear was found among the three genetic groups (data not shown). However, the proportion of extrathoracic involvement in group 1 (17.5%) was larger than those in group 2 (10.0%) and group 3 (13.1%).
Association between genetic grouping and site of disease.
The group 1 isolates occur more frequently in patients with extrathoracic TB than the group 2 isolates (14.1% versus 8.7%) (Table 3). When using group 2 as a referent, a significant association between infection by group 1 isolates and the extrathoracic involvement was found in a multivariate logistic regression analysis adjusting for age and HIV serostatus, race/ethnicity, sex, and plcD genotype (16, 28) (Table 4).
TABLE 3.
Frequency distribution of genetic groups, Beijing/W lineage, age, gender, race/ethnicity, HIV serostatus, and plcD genotype between the two sites of disease assessed by bivariate χ2 test or Fisher's exact test
| Variable | No. (%) positive for:
|
P value | |
|---|---|---|---|
| TTBa (n = 598) | ETTBb (n = 78) | ||
| Genetic group | 0.1685 | ||
| 2 | 394 (65.9) | 44 (56.4) | |
| 1 | 52 (8.7) | 11 (14.1) | |
| 3 | 152 (25.4) | 23 (29.5) | |
| Beijing/W lineagec | 0.0617 | ||
| No | 567 (95.1) | 70 (89.7) | |
| Yes | 29 (4.9) | 8 (10.3) | |
| Age (yr) | 0.3217 | ||
| <65 | 294 (49.2) | 43 (55.1) | |
| ≤65 | 304 (50.8) | 35 (44.9) | |
| Gender | 0.0013 | ||
| Male | 395 (66.1) | 37 (47.4) | |
| Female | 203 (33.9) | 41 (52.6) | |
| Race/ethnicity | 0.0013 | ||
| Non-Hispanic White | 350 (58.5) | 29 (37.2) | |
| Non-Hispanic Black | 193 (32.3) | 40 (51.3) | |
| Other | 55 (9.2) | 9 (11.5) | |
| HIV serostatus | 0.0004 | ||
| Negative | 276 (46.2) | 36 (16.2) | |
| Positive | 16 (2.7) | 9 (11.5) | |
| Unknown | 306 (51.2) | 33 (42.3) | |
| plcD genotype | 0.0293 | ||
| plcD wild type | 401 (67.1) | 44 (56.4) | |
| plcD mutation | 29 (4.8) | 9 (11.5) | |
| plcD and adjacent gene mutation | 168 (28.1) | 25 (32.1) | |
TTB, thoracic TB.
ETTB, extrathoracic TB.
Spoligotyping results were not available for two isolates, and, therefore, only 674 of the 676 isolates/patients were included in this analysis.
TABLE 4.
Associations of principal genetic groups and Beijing/W lineage with the site of diseasea
| Variable | Adjusted OR (95% CI) |
|---|---|
| Genetic group | |
| 2 | 1 |
| 1 | 2.40 (1.05, 5.52) |
| 3 | 1.24 (0.67, 2.31) |
| Beijing/W lineage | |
| No | 1 |
| Yes | 2.85 (1.33, 6.12) |
Associations were assessed by multivariate logistic regression models with GEE adjusting for potential confounding of age, gender, race/ethnicity, HIV serostatus, and plcD genotype.
Association between the Beijing/W lineage and site of disease.
We were concerned that the association between genetic group 1 and extrathoracic TB might be driven by the presence of the Beijing/W lineage, as this lineage is a subgroup of group 1. To address this concern, we performed a bivariate analysis and a multivariate logistic regression analysis separating group 1 into the Beijing/W lineage group 1 and the non-Beijing/W lineage group 1. The Beijing/W lineage in group 1 was found to be significantly associated with extrathoracic TB using group 2 as a referent in both bivariate (crude OR [95% CI], 2.46 [1.06, 5.72]) and multivariate analyses (adjusted OR [95% CI], 3.06 [1.39, 6.73]); however, the non-Beijing/W lineage group 1 isolate infection was not (crude OR [95% CI], 1.22 [0.35, 4.23]; adjusted OR [95% CI], 1.39 [0.29, 6.72]). We further compared the Beijing/W lineage-infected cases (n = 37) with all the non-Beijing lineage isolate-infected cases (n = 637) (Tables 3 and 4). In the multivariate logistic regression analysis, the Beijing/W lineage was significantly associated with extrathoracic TB in comparison with the non-Beijing/W lineage after controlling for age and other known risk factors for extrathoracic involvement (Table 4).
Association between genetic grouping and DNA fingerprint clustering.
Cases with isolates belonging to the same cluster, defined by a combination of IS6110 RFLP and pTBN12 secondary fingerprinting, are usually considered to be caused by TB transmissions that have occurred either at present or in the remote past (5, 11). We analyzed the frequency distribution of clustered strains among the three genetic groups with a sample consisting of 353 unique isolates and 91 isolates randomly selected from each of the 91 clusters identified during the study time period. In addition, in order to compare the results with those of a previous study (22), which defined a cluster as having more than five cases, we also conducted a similar analysis by studying only clusters having more than five cases. No statistically significant association was found between clustering and genetic grouping using either definition of clustering.
DISCUSSION
In order to increase our understanding of the clinical and epidemiological relevance of genetic grouping of M. tuberculosis clinical strains, we conducted a population-based study including analysis of 679 isolates from 679 patients diagnosed in Arkansas between 1 January 1996 and 31 December 2000. Patients infected by principal genetic group 1 isolates were more likely to have extrathoracic involvement than those infected by group 2 isolates. However, this association was driven by the association of infection by the Beijing/W lineage isolates, a subgroup of group 1, with extrathoracic involvement. In addition, we found that patients of Asian origin were the largest ethnic subgroup among group 1-infected patients, accounting for 42.9%; patients infected by the group 1 isolates were younger than those infected by the groups 2 and 3; and isolates in group 1 tended to have more genetic variations than those in groups 2 and 3 in terms of the range of the IS6110 copy number and the plc gene genotypes.
Although Beijing/W lineage strains have been hypothesized to be more virulent than non-Beijing/W lineage strains, few population-based studies that integrate molecular data with patient information have been conducted to date. Using a selected sample of 109 clinical isolates representing 33 strains of the Beijing genotype and 76 selected control strains of the non-Beijing genotypes from The Netherlands, Borgdorff et al. found no difference in chest X-ray presentations between infection by strains having the Beijing genotype and the infection by strains having the non-Beijing genotypes (4). Consistent with Borgdorff et al., we found similar pulmonary pathology between Beijing/W lineage isolate-infected individuals and those infected by the non-Beijing/W lineage isolates in terms of cavitation in lungs (44.8% versus 36.7%) and sputum smear positivity (50.0% versus 42.5%). However, we observed a statistically significant association (OR [95% CI], 2.85 [1.33, 6.12]) between the infection of Beijing/W lineage and extrathoracic TB, although the sample size of extrathoracic TB patients infected with Beijing/W lineage is relatively small (n = 8). The association between infection of the Beijing/W lineage isolates and extrathoracic TB remains to be further assessed using a study sample that contains a larger number of the extrathoracic TB patients infected with Beijing/W lineage isolates.
The overrepresentation of patients of Asian origin among patients infected with group 1 isolates is consistent with a previous report by Rhee and colleagues based on information from 419 United States-born TB patients diagnosed in San Francisco during the time period from 1991 through 1997 (21). However, in our study, the proportion of patients of Asian origin among the group 1 isolate-infected individuals was much higher than that found by the previous study (42.9% versus 14.8%). We found that 79% (27/34) of the patients of Asian origin were infected by an isolate belonging to group 1, which is twice as many as that found in San Francisco (9/22; 40.1%). One possible explanation is that the majority (33/34; 97.1%) of the Asian patients in our study were immigrants, whereas the San Francisco study only included the United States-born population. Our data support the previous notion that geographic separation might result in the establishment of a stable relationship between a distinct M. tuberculosis subpopulation and its host population (9, 10, 14).
Patients infected by the genetic group 1 isolates were younger than patients infected by isolates from groups 2 and 3. It was reported previously that the proportion of TB cases attributable to recent transmission is higher for young individuals than for the elderly (27). Thus, infections caused by isolates from group 1 are more likely to have resulted from recent transmissions than infections caused by isolates belonging to the other two groups. However, we did not observe an association between group 1 and genotyping-defined clustering. As clustering based on genotyping result could reflect both current and remote transmission (5, 11), Arkansas is a state with a stable and dispersed population, and the average age of TB patients in Arkansas is above the national average (5), it is possible that we could not differentiate clusters representing current transmission from those resulting from remote transmission. This kind of nondifferential misclassification tends to bias the result towards that of no association (23).
The group 1 isolates showed more genetic variations in the plc genes and a broader range of the IS6110 copy number than groups 2 and 3. This provides an additional support for the hypothesis that group 1 is evolutionarily older than groups 2 and 3 and therefore had more time to accumulate divergence (13, 22).
In conclusion, our results suggest that strains belonging to different genetic groups or lineages may have different potentials for virulence and pathogenicity. Clinical and epidemiological characterization of the major M. tuberculosis subpopulations circulating in different geographic regions is an essential step to developing better diagnostics, therapeutics, and vaccines for future TB control.
Acknowledgments
This study was supported by a grant from the National Institutes of Health (grant no. NIH-R01-AI151975).
We are indebted to Annadell H. Fowler, Don Cuningham, Bill Starrett, and Deborah Witonski for their valuable efforts that contributed to patient data and M. tuberculosis isolate collection and DNA fingerprinting of the study isolates. We acknowledge Kashef Ijaz's contribution to the establishment of the Arkansas Department of Health's surveillance database that was used for the study. We thank Dong Yang for her excellent technical assistance in the preparation of the genomic DNA of the study isolates and both Sarah Talarico and Dong Yang for their contribution to the collection of part of the genetic grouping data.
Footnotes
Published ahead of print on 13 December 2006.
REFERENCES
- 1.Barnes, P. F., and M. D. Cave. 2003. Molecular epidemiology of tuberculosis. N. Engl. J. Med. 349:1149-1156. [DOI] [PubMed] [Google Scholar]
- 2.Barnes, P. F., Z. Yang, S. Preston-Martin, J. M. Pogoda, B. E. Jones, M. Otaya, K. D. Eisenach, L. Knowles, S. Harvey, and M. D. Cave. 1997. Patterns of tuberculosis transmission in central Los Angeles. JAMA 278:1159-1163. [PubMed] [Google Scholar]
- 3.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45-52. [DOI] [PubMed] [Google Scholar]
- 4.Borgdorff, M. W., H. Van Deutekom, P. E. De Haas, K. Kremer, and D. Van Soolingen. 2004. Mycobacterium tuberculosis, Beijing genotype strains not associated with radiological presentation of pulmonary tuberculosis. Tuberculosis (Edinburgh) 84:337-340. [DOI] [PubMed] [Google Scholar]
- 5.Braden, C. R., G. L. Templeton, M. D. Cave, S. Valway, I. M. Onorato, K. G. Castro, D. Moers, Z. Yang, W. W. Stead, and J. H. Bates. 1997. Interpretation of restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates from a state with a large rural population. J. Infect. Dis. 175:1446-1452. [DOI] [PubMed] [Google Scholar]
- 6.Chaves, F., Z. Yang, H. el Hajj, M. Alonso, W. J. Burman, K. D. Eisenach, F. Dronda, J. H. Bates, and M. D. Cave. 1996. Usefulness of the secondary probe pTBN12 in DNA fingerprinting of Mycobacterium tuberculosis. J. Clin. Microbiol. 34:1118-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dale, J. W., G. H. Bothamley, F. Drobniewski, S. H. Gillespie, T. D. McHugh, and R. Pitman. 2005. Origins and properties of Mycobacterium tuberculosis isolates in London. J. Med. Microbiol. 54:575-582. [DOI] [PubMed] [Google Scholar]
- 8.Dolzani, L., M. Rosato, B. Sartori, E. Banfi, C. Lagatolla, M. Predominato, C. Fabris, E. Tonin, F. Gombac, and C. Monti-Bragadin. 2004. Mycobacterium tuberculosis isolates belonging to katG gyrA group 2 are associated with clustered cases of tuberculosis in Italian patients. J. Med. Microbiol. 53:155-159. [DOI] [PubMed] [Google Scholar]
- 9.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbon, M. Bobadilla del Valle, J. Fyfe, L. Garcia-Garcia, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. Leon, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendon, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188:759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gagneux, S., K. Deriemer, T. Van, M. Kato-Maeda, B. C. de Jong, S. Narayanan, M. Nicol, S. Niemann, K. Kremer, M. C. Gutierrez, M. Hilty, P. C. Hopewell, and P. M. Small. 2006. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 103:2869-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glynn, J. R., E. Vynnycky, and P. E. Fine. 1999. Influence of sampling on estimates of clustering and recent transmission of Mycobacterium tuberculosis derived from DNA fingerprinting techniques. Am. J. Epidemiol. 149:366-371. [DOI] [PubMed] [Google Scholar]
- 12.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutacker, M. M., J. C. Smoot, C. A. Migliaccio, S. M. Ricklefs, S. Hua, D. V. Cousins, E. A. Graviss, E. Shashkina, B. N. Kreiswirth, and J. M. Musser. 2002. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics 162:1533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirsh, A. E., A. G. Tsolaki, K. DeRiemer, M. W. Feldman, and P. M. Small. 2004. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc. Natl. Acad. Sci. USA 101:4871-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato-Maeda, M., J. T. Rhee, T. R. Gingeras, H. Salamon, J. Drenkow, N. Smittipat, and P. M. Small. 2001. Comparing genomes within the species Mycobacterium tuberculosis. Genome Res. 11:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong, Y., M. D. Cave, D. Yang, L. Zhang, C. F. Marrs, B. Foxman, J. H. Bates, F. Wilson, L. N. Mukasa, and Z. H. Yang. 2005. Distribution of insertion- and deletion-associated genetic polymorphisms among four Mycobacterium tuberculosis phospholipase C genes and associations with extrathoracic tuberculosis: a population-based study. J. Clin. Microbiol. 43:6048-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kremer, K., J. R. Glynn, T. Lillebaek, S. Niemann, N. E. Kurepina, B. N. Kreiswirth, P. J. Bifani, and D. van Soolingen. 2004. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J. Clin. Microbiol. 42:4040-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang, K., and S. Zeger. 1986. Longitudinal data analysis using generalized linear models. Biometrika 73:13-22. [Google Scholar]
- 19.Lopez, B., D. Aguilar, H. Orozco, M. Burger, C. Espitia, V. Ritacco, L. Barrera, K. Kremer, R. Hernandez-Pando, K. Huygen, and D. van Soolingen. 2003. A marked difference in pathogenesis and immune response induced by different Mycobacterium tuberculosis genotypes. Clin. Exp. Immunol. 133: 30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray, M. G., and W. F. Thompson. 1980. Rapid isolation of high molecular-weight plant DNA. Nucleic Acids Res. 8:4321-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhee, J. T., A. S. Piatek, P. M. Small, L. M. Harris, S. V. Chaparro, F. R. Kramer, and D. Alland. 1999. Molecular epidemiologic evaluation of transmissibility and virulence of Mycobacterium tuberculosis. J. Clin. Microbiol. 37:1764-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szklo, M., and F. J. Nieto. 2000. Part III. Threats to validity and issues of interpretation. Chapter 4. Understanding lack of validity: bias, p. 125-176. In M. a. N. Szklo and F. Javier (ed.), Epidemiology: beyond the basics. Aspen Publications, Gaitherburg, MD.
- 24.Talarico, S., R. Durmaz, and Z. Yang. 2005. Insertion- and deletion-associated genetic diversity of Mycobacterium tuberculosis phospholipase C-encoding genes among 106 clinical isolates from Turkey. J. Clin. Microbiol. 43:533-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Crevel, R., R. H. Nelwan, W. de Lenne, Y. Veeraragu, A. G. van der Zanden, Z. Amin, J. W. van der Meer, and D. van Soolingen. 2001. Mycobacterium tuberculosis Beijing genotype strains associated with febrile response to treatment. Emerg. Infect. Dis. 7:880-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vynnycky, E., N. Nagelkerke, M. W. Borgdorff, D. van Soolingen, J. D. van Embden, and P. E. Fine. 2001. The effect of age and study duration on the relationship between ‘clustering’ of DNA fingerprint patterns and the proportion of tuberculosis disease attributable to recent transmission. Epidemiol. Infect. 126:43-62. [PMC free article] [PubMed] [Google Scholar]
- 28.Yang, Z., D. Yang, Y. Kong, L. Zhang, C. F. Marrs, B. Foxman, J. H. Bates, F. Wilson, and M. D. Cave. 2005. Clinical relevance of Mycobacterium tuberculosis plcD gene mutations. Am. J. Respir. Crit. Care Med. 171:1436-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeger, S. L., and K. Y. Liang. 1986. Longitudinal data analysis for discrete and continuous outcomes. Biometrics 42:121-130. [PubMed] [Google Scholar]