Abstract
Strain subtyping is an important tool for detection of outbreaks caused by Salmonella enterica serotype Enteritidis. Current subtyping methods, however, yield less than optimal subtype discrimination. In this study, we describe the development and evaluation of a multiple-locus variable-number tandem repeat analysis (MLVA) method for subtyping Salmonella serotype Enteritidis. The discrimination ability and epidemiological concordance of MLVA were compared with those of pulsed-field gel electrophoresis (PFGE) and phage typing. MLVA provided greater discrimination among non-epidemiologically linked isolates than did PFGE or phage typing. Epidemiologic concordance was evaluated by typing 40 isolates from four food-borne disease outbreaks. MLVA, PFGE, and, to a lesser extent, phage typing exhibited consistent subtypes within an outbreak. MLVA was better able to differentiate isolates between the individual outbreaks than either PFGE or phage typing. The reproducibility of MLVA was evaluated by subtyping sequential isolates from an infected individual and by testing isolates following multiple passages and freeze-thaw cycles. PFGE and MLVA patterns were reproducible for isolates that were frozen and passaged multiple times. However, 2 of 12 sequential isolates obtained from an individual over the course of 36 days had an MLVA type that differed at one locus and one isolate had a different phage type. Overall, MLVA typing of Salmonella serotype Enteritidis had enhanced resolution, good reproducibility, and good epidemiological concordance. These results indicate that MLVA may be a useful tool for detection and investigation of outbreaks caused by Salmonella serotype Enteritidis.
Salmonella bacteria are a major source of human illness, causing an estimated 1.4 million annual cases of illness in the United States (29). Salmonella enterica serotype Enteritidis is the second most common serotype of Salmonella in the United States and among human clinical isolates in Europe (34). During 1985 to 1999, Salmonella serotype Enteritidis caused 29,762 illnesses, 2,904 hospitalizations, and 79 deaths in the United States (34).
Strain subtyping by molecular methods is a powerful tool for surveillance and outbreak investigation (3). Subtyping for surveillance and investigation of food-borne disease outbreaks attributable to Salmonella serotype Enteritidis, however, has been hampered by the fact that Salmonella serotype Enteritidis is one of the most genetically homogenous serotypes of Salmonella and is poorly differentiated by the most commonly used subtyping methods. Phage typing (PT) is a classical method traditionally used for subtype determination of Salmonella serotype Enteritidis but has limited discriminatory power and requires specialized phage collections that are available to only a few reference laboratories (20, 38, 39, 49). Plasmid profiling, single-enzyme ribotyping, and random amplified polymorphic DNA (RAPD) analysis also have limited discriminatory power for Salmonella serotype Enteritidis and, in addition, suffer from poor reproducibility (14, 20, 21, 27, 38, 39, 49). Two-enzyme ribotyping (PstI-SphI) may provide higher discriminatory power than other methods (6); however, manual ribotyping is labor-intensive while automated ribotyping is prohibitively expensive. Amplified fragment length polymorphism appears to be one of the more discriminatory methods for subtyping Salmonella serotype Enteritidis, but amplified fragment length polymorphism used with epidemiologically linked outbreak isolates of Salmonella serotype Enteritidis has revealed a lack of subtype stability (8, 42). In addition to limited resolution, each of these methods can be difficult to standardize and interlaboratory comparison is difficult. Pulsed-field gel electrophoresis (PFGE) is currently the gold standard for subtyping of Salmonella serotype Enteritidis, but PFGE also exhibits limited discriminatory power. Two PFGE patterns make up nearly 48% of the Salmonella serotype Enteritidis isolates in the PulseNet national database (Centers for Disease Control and Prevention [CDC], personal communication). Despite these drawbacks, PFGE has been used successfully to identify food-borne disease outbreaks (5). Because of the prevalence of Salmonella serotype Enteritidis as a pathogen and the importance of molecular subtyping in food-borne disease surveillance, there remains a need for a more suitable subtyping method that may be used with PulseNet to enable more timely detection of clusters and outbreaks.
Multiple-locus variable-number tandem repeat analysis (MLVA) is a subtyping technique that involves amplification and fragment size analysis of polymorphic regions of DNA containing variable numbers of tandemly repeated sequences. MLVA has been used to subtype a variety of species of bacteria, some of which have been difficult to subtype by other methods. Bacteria that have been typed by MLVA include Salmonella serotypes Typhimurium (22, 24, 35) and Typhi (26), Yersinia pestis (19), Francisella tularensis (9, 17), Mycobacterium tuberculosis (1), Haemophilus influenzae (41), Bacillus anthracis (18), Bordetella pertussis (40), Escherichia coli O157:H7 (23, 31), Enterococcus faecalis (47), Neisseria meningitidis (51), and Pseudomonas aeruginosa (33). MLVA has proven to be a rapid method and may be easy to standardize between laboratories. This study describes the development of an MLVA typing scheme and comparison of MLVA, PT, and PFGE for subtyping of Salmonella serotype Enteritidis.
MATERIALS AND METHODS
Bacterial strains.
One hundred fifty-three Salmonella serotype Enteritidis isolates recovered from Minnesota residents during the years 1998 to 2003 were selected for use in development of an MLVA typing scheme and for comparing the discriminatory power and epidemiologic concordance of PT, PFGE, and MLVA. Included in the 153 isolates were 40 isolates from four separate food-borne disease outbreaks and 113 isolates without any known epidemiological links (sporadic isolates). Outbreak isolates were from well-characterized outbreaks that contained more than eight cases and for which a common source was found. Cases were interviewed regarding their demographic information, recent travel, and food consumption prior to becoming ill. Nine of the sporadic cases could not be interviewed. An additional 12 Salmonella serotype Enteritidis isolates cultured sequentially from a single individual were used to evaluate the stability of the loci chosen for inclusion in the MLVA typing scheme.
Identification of variable-number tandem repeats (VNTRs).
Tandem repeat finder (TRF) software (2) was used to identify tandem repeats in the partial genomic sequence of Salmonella serotype Enteritidis phage type 8 (strain LK5; University of Illinois-Urbana-Champagne, www.salmonella.org) and the completed genomic sequence of Salmonella serotype Enteritidis phage type 4 (strain PT4; Sanger Institute, www.sanger.ac.uk/Projects/Salmonella). Because of the relative lack of genomic sequence data available for Salmonella serotype Enteritidis, the genomic sequences of S. enterica serovar Typhi strains Ty2 and CT18 and Salmonella serotype Typhimurium strain LT2 were also screened using TRF in hopes of finding additional candidate repeat loci that may also be present in Salmonella serotype Enteritidis. To screen for variability in the number of tandem repeats, PCR primers targeting the regions flanking tandem repeat loci identified by TRF were designed. These primers were used to amplify DNA from a set of 12 genetically diverse Salmonella serotype Enteritidis isolates as determined by PFGE and/or PT.
Polymorphism detection by RAPD.
Because of the paucity of sequence data available, RAPD was used to screen Salmonella serotype Enteritidis for regions of polymorphic DNA that may contain tandem repeats. DNA was extracted from Salmonella serotype Enteritidis strains with the common PFGE type SE1B1 using a QIAGEN DNA Mini Kit (QIAGEN, Valencia, CA). PCR for RAPD analysis was performed as described previously (21) except that the 72°C terminal extension was lengthened to 10 min. PCR products were separated on a 2% agarose gel. Polymorphic fragments were excised from the gel, purified using a QIAquick spin column (QIAGEN), and cloned using a Topo TA cloning kit (Invitrogen, Carlsbad, CA). Cloned fragments were amplified from the cloning vector by PCR using M13 forward (−20) and M13 reverse primers and sequenced on a Beckman-Coulter CEQ 8000 automated DNA sequencer using a Dye Terminator Cycle Sequencing Quick Start kit (Beckman-Coulter, Fullerton, CA).
MLVA typing.
DNA was extracted from Salmonella serotype Enteritidis isolates grown overnight on MacConkey agar plates by suspending a small loopful of cells in 200 μl DNAzol (Molecular Research Center, Inc., Cincinnati, OH) and boiling the suspension for 5 min. Following boiling, the sample was centrifuged and the supernatant applied to a QIAquick PCR purification kit (QIAGEN), washed, and eluted into 50 μl elution buffer following the manufacturer's directions.
The 10 loci ultimately selected for MLVA were amplified individually by PCR. Reaction mixtures contained 1× QIAGEN PCR buffer (containing 1.5 mM MgCl2), 0.2 mM deoxynucleoside triphosphates, 1 μM forward primer labeled with a WellRED-labeled dye (Proligo, Boulder, CO), 1 μM nonlabeled reverse primer, 0.6 U HotStarTaq DNA polymerase (QIAGEN), 2.5 μl template DNA diluted 1:100, and sterile water to a volume of 25 μl. The PCR conditions were 95°C for 15 min, cycling at 94°C for 1 min, the specific annealing temperature for each locus (Table 1) for 1 min, and 72°C for 1 min for 35 cycles, with a final incubation of 72°C for 10 min. The fragment size for each locus, with the exception of SE-7, was determined by combining 1.0 to 2.0 μl PCR product, 37.5 μl formamide, and 0.5 μl 600-bp DNA size standard (Beckman-Coulter) followed by fragment size analysis using a Beckman-Coulter CEQ 8000 automated DNA sequencer. The following conditions were used for separation of fragments for all loci except SE-7: capillary temperature of 35°C and denaturation temperature of 90°C, duration of 120 seconds; injection voltage of 2.0 kV, duration of 30 seconds; separation voltage of 7.5 kV, duration of 50 min. The fragment size for SE-7 was determined using the same conditions as for the other loci with the following exceptions: a 25- to 1,000-bp DNA size standard (BioVentures, Murfreesboro, TN), a capillary temperature of 50°C, a separation voltage of 5.0 kV, and a separation duration of 80 min.
TABLE 1.
Primers and annealing temperatures used for MLVA
| Locus | Primer | Primer sequence | Annealing temp (°C) |
|---|---|---|---|
| SE-1a | SE-1F | 5′-AGACGTGGCAAGGAACAGTAG-3′ | 64 |
| SE-1R | 5′-CCAGCCATCCATACCAAGAC-3′ | ||
| SE-2a | SE-2F | 5′-CTTCGGATTATACCTGGATTG-3′ | 58 |
| SE-2R | 5′-TGGACGGAGGCGATAG-3′ | ||
| SE-3a | SE-3F | 5′-CAACAAAACAACAGCAGCAT-3′ | 58 |
| SE-3R | 5′-GGGAAACGGTAATCAGAAAGT-3′ | ||
| SE-4b | SE-4F | 5′-ACTTTAGAAAATGCGTTGAC-3′ | 50 |
| SE-4R | 5′-AAGTCAACTGCTCTACCAAC-3′ | ||
| SE-5b | SE-5F | 5′-CGGGAAACCACCATCAC-3′ | 50 |
| SE-5R | 5′-CAGGCCGAATAGCAGGAT-3′ | ||
| SE-6b | SE-6F | 5′-CCCCTAAGCCCGATAATG-3′ | 50 |
| SE-6R | 5′-GCCGTTGCTGAAGGT-3′ | ||
| SE-7a | SE-7F | 5′-GATAATGCTGCCGTTGGTAA-3′ | 58 |
| SE-7R | 5′-ACTGCGTTTGGTTTCTTTTCT-3′ | ||
| SE-8c | SE-8F | 5′-TTGCCGCATAGCAGCAGAAGT-3′ | 50 |
| SE-8R | 5′-GCCTGAACACGCTTTTTAATAGGCT-3′ | ||
| SE-9c | SE-9F | 5′-CGTAGCCAATCAGATTCATCCCGCG-3′ | 45 |
| SE-9R | 5′-TTTGAAACGGGGTGTGGCGCTG-3′ | ||
| SE-10c | SE-10F | 5′-GCTGAGATCGCCAAGCAGATCGTCG-3′ | 45 |
| SE-10R | 5′-ACTGGCGCAACAGCAGCAGCAACAG-3′ |
Primer sequence derived from Salmonella serotype Enteritidis LK5 (phage type 8) partial genome sequence (http://www.sanger.ac.uk/Projects/Salmonella/SEN_genepred.emb).
Primer sequence derived from Salmonella serotype Typhimurium LT2 genome sequence (reference sequence number NC 003197).
Primer sequence derived from Salmonella serotype Enteritidis PT4 genome sequence (http://www.salmonella.org/genomics/sen.dbs).
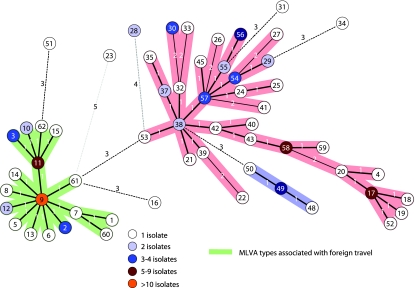
Following fragment analysis, each locus was assigned a variant score based on the fragment size. Each isolate was assigned an MLVA type based on compilation of the variant score of the 10 loci. Variant scores were entered into BioNumerics software (Applied-Maths, St.-Martens-Latem, Belgium) as categorical data. Cluster analysis of the categorical data was done by generation of either dendrograms or minimum spanning trees. Minimum spanning trees are frequently used to cluster subtyping data composed of categorical variables, such as multilocus sequence typing and MLVA, and are useful for visualizing the relationship between large numbers of isolates in a single, compact image. The distance between any pair of organisms is defined as the number of different characters, so the distance between isolates differing at one locus is 1 and at two loci is 2, etc. The minimum spanning tree is constructed so that the total summed distance along the branches is minimized. A detailed description of analysis of MLVA data using minimum spanning trees may be found in the study by Schouls et al. (40). The parameters used for generating the minimum spanning trees in BioNumerics are provided in the legend to Fig. 1.
FIG. 1.
Minimum spanning tree of MLVA of sporadic Salmonella serotype Enteritidis isolates. A categorical coefficient and the priority rule using the highest number of single-locus changes were used for generation of the minimum spanning tree. Each circle in the tree represents a different MLVA type, with the MLVA type number indicated by the number in the circle. Heavy short lines connecting two MLVA types denote types differing by a single MLVA locus while thin longer lines connect double-locus variants, and dotted lines indicate the most likely connection between two types differing by more than two MLVA loci. The number of loci that differ between two MLVA types is indicated on the lines connecting the MLVA types. The colors of the circles indicate the numbers of isolates with a particular MLVA type as indicated on the figure. Clusters were defined as MLVA types having a maximum distance of changes at two loci and a minimum cluster size of two types. Cluster A is shaded in green, cluster B in blue, and cluster C in pink.
PFGE and phage typing.
Salmonella serotype Enteritidis isolates were subtyped by PFGE using restriction endonucleases XbaI (Promega) and BlnI (Roche) according to published methods (37). PFGE patterns were compared on dendrograms generated in BioNumerics using the Dice coefficient and a 1% band matching criterion. Patterns with no noticeable differences were considered indistinguishable and were assigned the same PFGE pattern designation. The XbaI and BlnI pattern designations were combined to form a cumulative PFGE pattern designation. For example, an isolate with the XbaI pattern SE1 and the BlnI pattern B1 is designated as PFGE type SE1B1. Phage typing was performed according to standardized methods at the Centers for Disease Control and Prevention (13, 50).
Statistical analysis.
Diversity of VNTR variants for each locus was assessed using Nei's measure of allelic diversity (30). Diversity of MLVA, PFGE, and PT was assessed using Simpson's index of diversity (D) (15) and the Shannon-Weiner (Shannon) diversity (H′) and equitability (E) indices (43, 52) calculated using the Biodiversity Calculator developed by J. Danoff-Burg and C. Xu (http://www.columbia.edu/itc/cerc/danoff-burg/MBD_Links.html). For the Simpson indices, variances and confidence intervals (CIs) were calculated as described previously (12). Statistical comparisons of the Shannon and Simpson indices were performed by t test (28). Odds ratios (ORs) for the evaluation of risk factors with Salmonella serotype Enteritidis clusters were calculated using Epi Info version 6.04d (Centers for Disease Control and Prevention and World Health Organization, Geneva, Switzerland).
RESULTS
VNTR identification and analysis.
One hundred fifty-seven tandem repeat loci were evaluated using a panel of 12 isolates of Salmonella serotype Enteritidis previously shown by PFGE and PT to be genetically diverse. Ten tandem repeat loci exhibited adequate diversity in the screening panel and were selected for evaluation as part of an MLVA typing system (Table 2). Repeat-containing loci were identified in both coding and intergenic regions of the chromosome. The tandem repeat units in loci selected for use in the MLVA subtyping scheme varied in size from 6 to 117 bp, and the number of variants ranged from 2 to 11, depending on the locus. Nei's diversity index (DI) was calculated for individual loci based on the 57 different MLVA scores obtained from testing of sporadic and outbreak isolates of Salmonella serotype Enteritidis and ranged from 0.05 to 0.77 (Table 2). All loci demonstrating the ability to discriminate between sporadic isolates, particularly of common PFGE subtype SE1B1, were included in further analyses regardless of diversity index. Nine of these 10 loci were identified by TRF analysis of the Salmonella serotype Enteritidis genomic sequences. Locus SE-8 was identified as a polymorphic band by RAPD analysis and was subsequently found to contain an 87-bp tandem repeat. This locus was found to be variable among Salmonella serotype Enteritidis isolates; however, the 87-bp repeat sequence was present in only a single copy in many Salmonella serotype Enteritidis isolates, including the sequenced strains LK5 and PT4. This explains why this locus was not identified by TRF analysis using those sequences. The loci designated SE-5 and SE-6 are the same as those analyzed in previous Salmonella MLVA publications (22, 24, 35).
TABLE 2.
Characteristics of loci used for MLVA
| Locus | Repeat size (bp) | Repeat no. in straina:
|
No. of variantsb | Nei's diversity indexc | Coding region | |
|---|---|---|---|---|---|---|
| LK5 | PT4 | |||||
| SE-1 | 7 | 4.9 | 4.9 | 6 | 0.62 | None identified |
| SE-2 | 7 | 4.7 | 5.7 | 8 | 0.73 | None identified |
| SE-3 | 12 | 4.0 | 3.0 | 3d | 0.54 | None identified |
| SE-4 | 117 | 2.7 | 2.7 | 3 | 0.05 | valU |
| SE-5 | 6 | NF | 11.8 | 11 | 0.77 | yohM |
| SE-6 | 33 | 10.8 | 10.8 | 4 | 0.07 | bigA |
| SE-7 | 61 | 8.5 | 8.5 | 7d | 0.60 | ygbF |
| SE-8e | 87 | 1 | 1 | 2 | 0.45 | None identified |
| SE-9 | 9 | 2.1 | 3.1 | 4 | 0.53 | ushA |
| SE-10 | 45 | 3.9f | 8.1 | 2 | 0.05 | tolA |
Repeat number was determined from the genomic sequence using TRF, which identified partial repeats in flanking sequences. Inspection of loci SE-2 and SE-9 in the sequences of PT4 and LK5 revealed that they vary by one whole repeat unit. NF indicates that the sequence was not found in the incomplete sequence of LK5.
Number of different fragment size polymorphisms detected among 156 Salmonella serotype Enteritidis isolates tested, including outbreak and sporadic isolates.
1 − ∑(variant frequency). See footnote b.
Includes a null variant (no fragment amplified by PCR).
Locus SE-8 was identified by RAPD marker analysis and was found to be variable in strains of Salmonella serotype Enteritidis other than LK5 and PT4.
Partial sequence.
Stability.
To analyze the in vivo stability of VNTR loci, we analyzed isolates from 12 stool samples from a single person submitted to the Minnesota Department of Health over a 36-day period. MLVA, PFGE, and PT were performed on single colonies from frozen isolate stocks for each sample. Nine of the loci were stable for all isolates, and two of the samples yielded differences at a single locus, SE-2. The fragment sizes for SE-2 loci of the different MLVA types were consistent with the loss of two repeats for isolate 8 and addition of one repeat for isolate 12, compared to the predominant MLVA type. Isolate 1 was phage type 34a while the other 11 isolates were phage type 13a; however, the PFGE patterns were consistent for all 12 isolates.
In order to analyze the in vitro stability of the VNTR loci, we performed multiple subcultures and multiple freeze-thaw cycles on single-colony isolates of two genetically diverse strains of Salmonella serotype Enteritidis (MLVA types MSE30 and MSE9). Five individual colonies of each strain were picked to determine homogeneity of the initial plate. The isolates were passaged every day for 10 days, and 10 single colonies were picked from the final subculture plate and subjected to MLVA typing. To analyze the effect of frozen storage on VNTR stability, a sweep of Salmonella serotype Enteritidis from the original plate was frozen in glycerol at −80°C overnight. The following day, the isolate was grown on a tryptic soy broth agar plate overnight at 37°C. The isolate was frozen and cultured four more times. Following the fifth freeze-thaw cycle, 10 colonies were picked for MLVA. All picks from the original plate, the multiple subcultures, and the freeze-thawed samples exhibited the same MLVA type as the original isolate (data not shown).
Diversity of MLVA, PFGE, and phage types among sporadic isolates.
One hundred thirteen sporadic isolates were analyzed by MLVA, PFGE, and PT. There were 57 MLVA types, 33 PFGE types, and 15 phage types among the sporadic isolates (Table 3). Seven of the 113 isolates yielded an indeterminate phage type (reactive but did not conform). Although there were twice as many subtypes identified by PFGE as by PT, the Simpson diversity indices calculated for PFGE and for PT were similar, the confidence intervals overlapped, and the differences were not statistically significant (P = 0.7). The distributions of PFGE and phage types were not even. Fifty-six percent of the sporadic isolates were assigned to only two PFGE types, SE1B1 (n = 34) and SE11B6 (n = 29), and 64% of the sporadic isolates fell into three phage types, PT4 (n = 29), phage type 13a (n = 26), and phage type 8 (n = 17) (Table 3). Because Simpson's diversity index did not reveal a difference in discriminatory power between PFGE and PT despite the fact that PFGE subtyping yielded twice as many subtypes as did PT, we also calculated Shannon's diversity index for each method. Shannon's index calculates separate values for the diversity index (H′), which is an indicator of species richness (i.e., number of subtypes), and an equitability index (E), which is a measure of the evenness of subtype distribution. Shannon's H′ was slightly higher for PFGE than for PT (P = 0.071) and better reflected the larger number of subtypes produced by PFGE. However, the value E was found to be higher for PT than for PFGE, presumably due to the slightly more even distribution of phage types. Both the Simpson and Shannon diversity indices for MLVA were significantly higher (P < 0.001) than those for PFGE and PT, revealing that MLVA provided both a greater number of different subtypes and a more even distribution. Isolates with the common PFGE types SE11B6 and SE1B1 were divided into 12 and 19 MLVA types, respectively. Isolates with the common phage types 4, 13a, and 8 were divided into 14, 17, and 11 MLVA types, respectively. The distribution of MLVA types was more equitable than that of either PFGE or phage types as reflected by Shannon's E value, with only 20% of the isolates belonging to the two most common MLVA types (MSE9 and MSE11). However, these common MLVA types could be further discriminated by both PFGE and phage typing. MLVA type MSE11 contained four PFGE types and five phage types while MLVA type MSE9 contained five PFGE types and four phage types. These data indicate that, regardless of the primary subtyping method used, a secondary method may be needed to resolve common Salmonella serotype Enteritidis subtypes.
TABLE 3.
Comparison of typing methods in sporadic isolates (n = 113)
| Method | No. of types | Frequent type (n) | Diversity indexc
|
|
|---|---|---|---|---|
| Simpson's (CI) | Shannon'sa | |||
| PFGE | 33 | SE1B1 (34) | 0.839 (0.791-0.887) | H′, 2.465 |
| SE11B6 (29) | E, 0.705 | |||
| MLVA | 57 | MSE9 (17) | 0.965 (0.947-0.983) | H′, 3.661 |
| MSE11 (6) | E, 0.902 | |||
| PT | 15b | Phage type 4 (29) | 0.850 (0.815-0.885) | H′, 2.177 |
| Phage type 13a (26) | E, 0.7852 | |||
| Phage type 8 (17) | ||||
Shannon-Weiner diversity index where H′ represents the subtype diversity, i.e., the number of different subtypes, and E is a measure of evenness, i.e., how evenly the subtypes are distributed in the population sampled.
Seven isolates were reactive but did not conform to a known pattern, and these isolates were considered to be untypeable.
P values calculated for PFGE and PT compared to MLVA were <0.01 for both diversity indices. P values for PFGE compared to PT were >0.5 for Simpson's DI and <0.05 for Shannon's DI.
Cluster analysis.
BioNumerics was used to generate a minimum spanning tree of MLVA types of sporadic isolates of Salmonella serotype Enteritidis (Fig. 1). Minimum spanning trees illustrate the relationship of subtypes based on the number of VNTR loci that differ between two MLVA profiles. Because low-diversity loci may provide phylogenetic information (44), all low-diversity loci, with the exception of SE-4, which has no discernible effect on clustering, were used for creation of the minimum spanning tree. Evaluation of the contribution of the individual MLVA loci to clustering of isolates in the minimum spanning tree indicated that elimination of low-diversity loci (in particular SE-6 and SE-10) had a negligible effect on the discriminatory power of MLVA but reduced the complexity of the branching of the minimum spanning tree (data not shown).
The minimum spanning tree analysis revealed three clusters of MLVA types that differ by two or more loci from all other MLVA types. Statistical analysis of the epidemiology data on individuals whose isolates comprised MLVA group A (shaded green in Fig. 1) revealed that these individuals were significantly more likely to have traveled outside of the United States in the 7 days before the onset of illness than were individuals whose isolates did not belong to group A. Thirty-four of 42 cases in group A traveled outside the United States shortly before becoming ill compared to 3 of 62 non-group A cases (OR, 84; 95% CI, 19 to 483; P < 0.001). Of the 34 cases in group A who traveled outside of the United States shortly before becoming ill, 19 traveled to South America, 8 traveled to Asia, 6 traveled to Europe, and 1 traveled to Africa. Minimum spanning trees cannot be performed on PFGE patterns; however, clusters from PFGE dendrograms derived from XbaI (OR, 23; 95% CI, 7 to 76; P < 0.001), BlnI (OR, 38; 95% CI, 11 to 142; P < 0.001), and an XbaI-BlnI composite (OR, 47; 95% CI, 13 to 191; P < 0.001) also reveal a correlation with foreign travel. The majority of the isolates in cluster A were phage type 4 (n = 25). Of the 29 cases whose isolates were phage type 4, 17 reported a history of foreign travel, and three were not interviewed. Four additional isolates with phage type 4 were not included in cluster A, and three of these were in cluster B (blue shaded). Cases with these four phage type 4 isolates did not report a history of foreign travel.
Epidemiologic concordance.
Subtype analysis of Salmonella serotype Enteritidis isolates from four food-borne outbreaks showed that all isolates within an outbreak were indistinguishable by each of the methods, with the exception of a single isolate from outbreak 2 that had a different phage type than that of the other 10 isolates from that outbreak (Table 4). Evaluation of isolate subtypes between outbreaks revealed that, although a different MLVA type was associated with each outbreak, two of the outbreaks were associated with a single PFGE type, and all four outbreaks were associated with a single phage type. Had outbreaks 3 and 4 occurred simultaneously, MLVA would have been the only method able to accurately discriminate isolates from the two outbreaks. The PFGE patterns for strains linked to outbreaks 1 and 2 were uncommon among the sporadic isolates. However, strains involved in outbreaks 3 and 4 had the most common PFGE pattern, SE1B1, which was present in 30% of the sporadic isolates. Isolates from all four outbreaks were phage type 13a (with the exception of one isolate), which is one of the most common phage types in the United States and was present in 23% of the sporadic isolates tested. In contrast to PFGE and PT, the MLVA patterns for outbreaks 1, 2, and 3 were seen in four (3.5%) of the sporadic isolates. The MLVA type of the strains from outbreak 4, MSE58, was seen in seven sporadic isolates (6.2%). However, six of the sporadic isolates with MLVA type MSE58 were seen within 2 weeks of outbreak 4 and had the same PFGE type and phage type as the outbreak isolates did. These data suggest that there may have been an unidentified link between these sporadic cases with MSE58 and those associated with outbreak 4.
TABLE 4.
Comparison of methods for outbreak investigation
| Outbreak (n) | Pattern of outbreak type (no.)
|
Occurrence of outbreak type among sporadic isolates [no. (%)]
|
||||
|---|---|---|---|---|---|---|
| PFGE | PT | MLVA | PFGE | PT | MLVA | |
| OB 1 (8) | SE77B52 | 13a | MSE32 | 1 (0.9) | 26 (23) | 1 (0.9) |
| OB 2 (11) | SE82B1 | 13a (10) | MSE46 | 0 (0) | 26 (23) | 0 (0) |
| RDNCb (1) | 7 (6.2) | |||||
| OB 3 (11) | SE1B1 | 13aa | MSE54 | 34 (30) | 26 (23) | 3 (2.7) |
| OB 4 (10) | SE1B1 | 13a | MSE58 | 34 (30) | 26 (23) | 7 (6.2) |
Only three isolates were phage typed.
RDNC, reactive but does not conform to known pattern.
DISCUSSION
Detection of food-borne outbreaks due to Salmonella serotype Enteritidis relies on a suitable method for subtype determination. Although standards for subtyping methods do not exist, the European Study Group for Epidemiological Markers recommended that typing methods meet certain criteria, including discriminatory power, stability, epidemiologic concordance, and convenience criteria, such as rapidity and ease of use (45). More recently, the Clinical and Laboratory Standards Institute (CLSI) has issued a proposed guideline for characterizing and validating bacterial typing systems (7). Current Salmonella serotype Enteritidis subtyping methods for the most part do not meet the proposed criteria, with the main problem being suboptimal discriminatory power. The method of choice for the national PulseNet surveillance system has been PFGE, although ribotyping may provide higher discriminatory power than does PFGE with a single enzyme (6). Ribotyping is not widely used for Salmonella serotype Enteritidis subtyping in the United States, however, and suffers from the many of the same data portability issues as PFGE does. In an attempt to address the issues of discriminatory power and data portability, we developed and evaluated an MLVA method for subtyping Salmonella serotype Enteritidis.
Discriminatory power, defined as the ability to distinguish between unrelated strains, is commonly used to evaluate subtyping methods. An objective measure of discriminatory power may be obtained by calculation of a DI. Diversity is a function of species (or subtype) richness, which refers to the number of subtypes in a population, and subtype evenness, which refers to the relative distribution, or abundance, of individuals among the different subtypes. A number of statistical formulas are available to calculate diversity indices, and each places a slightly different emphasis on subtype richness and evenness. Simpson's DI is the mathematical measure most commonly used as an estimate of the discriminatory ability of subtyping methods. One reason for the popularity of the Simpson DI is that it has biological meaning, i.e., it is a measure of the probability that two epidemiologically unrelated isolates will be characterized as being “different” by the typing method being evaluated. Simpson's DI is heavily weighted towards the relative abundance of each subtype in the population studied. Simpson's DI is less sensitive to subtype richness. As calculated by Simpson's formula, a subtyping method that identified three subtypes distributed equally among the population would receive a higher score than a method that identified six subtypes but whose distribution in the population was unequal. In contrast, the Shannon-Weiner, or Shannon, diversity index is more influenced by the number of species. Shannon's formula calculates separate indices for diversity and for evenness. In this study, we calculated diversity indices for all three typing methods using Shannon's and Simpson's formulas. While Simpson's index indicated that MLVA exhibited the highest discriminatory power, PFGE and PT were found to be essentially equivalent in spite of the greater number of subtypes obtained by PFGE subtyping due to Simpson's heavy emphasis on the evenness of subtype distribution. Shannon's index also indicated that MLVA was more discriminatory than either PFGE or PT; however, unlike Simpson's index, Shannon's index indicated that PFGE was more discriminatory than PT, which is more consistent with the greater number of subtypes and better epidemiologic concordance of PFGE than of PT. The value of Shannon's E for phage typing was greater than that of PFGE, however, reflecting the more even distribution of phage types than of PFGE patterns. These results indicate that Shannon's index, although not as easily interpreted as Simpson's index, may provide a better objective measure of the performance characteristics of a subtyping method.
Regardless of the diversity index used, MLVA appears to provide improved discriminatory power for subtyping. However, some clonality is still observed among strains typed by MLVA; therefore, a combination of typing methods may be required in instances where greater discriminatory power is needed. BioNumerics software allows for the creation of composite data sets consisting of multiple different data types, which may include character data, gels, and sequence information. Composite data sets may be analyzed in the same manner as single data types, and the use of combined Salmonella serotype Enteritidis subtype methods in a composite data set should result in enhanced discriminatory power.
While measures of discriminatory power are useful in the evaluation of a subtyping method, epidemiologic concordance is equally, if not more, important for evaluation of the utility of the typing method. We define epidemiologic concordance as the ability of a subtyping method to (i) correctly identify epidemiologically linked isolates as being genetically related, (ii) differentiate epidemiologically linked isolates from background sporadic isolates, and (iii) differentiate between outbreaks not due to a common source. Evaluation of Salmonella serotype Enteritidis isolates from four epidemiologically well-characterized food-borne outbreaks indicated that both MLVA typing and PFGE correctly clustered isolates within an outbreak, i.e., all isolates within a defined outbreak had the same pattern. MLVA provided better discrimination of isolates between outbreaks than did either PFGE or PT and provided better discrimination between strains with outbreak patterns and sporadic isolates. These results indicate that MLVA, while yielding improved discriminatory power, also provides sufficient epidemiologic concordance to be useful in outbreak investigations.
Clustering can provide useful information on strains that are related. Minimum spanning tree analysis of MLVA subtypes revealed a cluster of isolates that were found to be associated with foreign travel. Clusters of isolates identified from dendrograms of PFGE restriction patterns also revealed a correlation with foreign travel, and while statistically significant, the correlation was not as strong as that seen with the MLVA minimum spanning tree. Of the 44 isolates in the MLVA cluster, 25 of them were phage type 4 and 6 were phage type 1. Five isolates within the cluster could not be assigned a phage type, but the cases associated with these isolates reported a history of recent foreign travel. Clustering of Salmonella serotype Enteritidis isolates associated with foreign travel may be a reflection of the different predominant Salmonella serotype Enteritidis phage types seen throughout the world. Phage type 4 emerged in Europe in the early 1990s and is now the predominant phage type in much of the world, including Europe (32), Singapore (25), Brazil (10), Chile (11), and Thailand (4). Phage type 4 is relatively uncommon in parts of the United States and Canada; however, it has been found in areas of the West Coast of the United States and has been associated with foreign travel in people returning to the United States and Canada (16, 29a). Eight of the 42 patients in MLVA group A did not report foreign travel before becoming ill, and four patients had phage type 4 strains that did not cluster with group A, nor did they report a history of foreign travel. These patients may have acquired their infections from secondary transmission or by consuming contaminated imported food, or the infections may have been domestically acquired.
VNTR loci have been found to differ with respect to stability and have been found to exhibit variability during persistent colonization of an individual (36, 48). CLSI has recommended that the “biological reproducibility” of a bacterial strain typing method be assessed by analysis of epidemiologically linked (i.e., outbreak-related) sets of isolates and isolates from a single episode of infection in an individual (7). MLVA typing of Salmonella serotype Enteritidis isolates following successive culturing and freeze-thaw cycles showed that VNTR loci remained stable and the MLVA patterns did not change, at least in the two isolates tested in vitro. In addition, we have not observed any variability among VNTR fragment sizes from isolates repeatedly subcultured and used as control strains. MLVA of 12 Salmonella serotype Enteritidis isolates recovered from a single individual over a 36-day period, however, revealed variability at one locus, SE-2. The PFGE pattern of these isolates remained stable, but one isolate had a different phage type. These data suggest that the selective pressures on isolates in vivo may be greater than the pressures that occur in vitro. MLVA patterns of 40 isolates from four outbreaks used to evaluate epidemiologic concordance showed that MLVA patterns from epidemiologically linked outbreak isolates remained stable. Taken together, these data indicate that these VNTR loci exhibit sufficient stability for use in characterizing outbreaks. It remains possible that patterns will exhibit some variability if there is secondary transmission or if individuals are colonized for some period prior to sampling. Additional data will be necessary to adequately characterize the stability of these VNTR loci. Interpretive criteria that account for genetic variability of MLVA patterns analogous to the Tenover criteria used for PFGE may need to be developed (46).
We chose to evaluate all isolates using all of the VNTR loci identified in our screening in order to make an accurate determination of the diversity of each locus in our sample set. Ultimately, though, the individual loci included in an MLVA protocol will depend on the application of the typing method and the nature of the strains being examined. For example, high-diversity loci may be helpful for differentiation of isolates from a geographically limited set of isolates, such as the ones that we examined in this study, while low-diversity loci have been shown to act as a molecular clock and may be particularly useful for phylogenetic analysis in isolates that are more geographically dispersed (44). In addition, certain VNTR loci have been shown to exhibit what has been termed “allele bias,” i.e., certain size variants or alleles are dominant, enabling these markers to be used to identify different biovars of F. tularensis (9). Our experience with subtyping of group B salmonellae using MLVA (unpublished data) indicates that allele bias is helpful in identification of different serovars. Because VNTR data are entered into BioNumerics as categorical data, individual loci may be readily included in or removed from the analysis. This enables the user to tailor the method as needed to obtain the desired level of discriminatory power. This flexibility would allow laboratories to use a standard set of loci to facilitate interlaboratory comparison of MLVA data, while providing the option to use additional loci to improve resolution or to determine phylogenetic relationships.
There were some limitations inherent in this study. Although the PFGE and phage types of Salmonella serotype Enteritidis isolates included in this study are representative of the most common PFGE and phage types in the United States, all of the isolates evaluated were from Minnesota residents. Evaluation of isolates with a more global distribution will be necessary to evaluate the suitability of MLVA as a global method for typing of Salmonella serotype Enteritidis. Another limitation is that the study was retrospective, and the outbreak isolates included in the evaluation were initially identified as temporal clusters of Salmonella serotype Enteritidis with the same PFGE type. This results in an inherent bias in favor of PFGE. While epidemiologic investigation supported a common exposure among case patients with these isolates, misclassification and failure to identify outbreaks among sporadic isolates cannot be ruled out. Nevertheless, in spite of this bias, MLVA was found to provide excellent epidemiologic concordance and superior discrimination between outbreaks compared with PFGE. Finally, while data portability is facilitated by the use of sequence-based subtyping methods, our use of fragment analysis to assess VNTR polymorphisms is subject to some of the same limitations seen for other gel-based systems. Improvements in DNA sequencing technology, for example the ability to obtain longer reads by using pyrosequencing, will enable more rapid and precise determination of VNTR polymorphisms.
Our data indicate that MLVA may be a viable alternative to PFGE for subtyping of Salmonella serotype Enteritidis. Ultimately, however, interlaboratory reproducibility and convenience factors including cost, ease of use, data portability, and turnaround time need to be assessed for MLVA, or any alternative subtyping method, prior to implementation by PulseNet.
Acknowledgments
This study was developed as part of a PulseNet USA initiative to develop and evaluate new subtyping methods. This publication was supported by grant cooperative agreement number U60-CCU303019 from CDC and APHL for development and validation of DNA sequence-based methods for subtyping of Salmonella for PulseNet.
Its contents are solely the responsibility of the authors and do not necessarily represent the official views of APHL or CDC.
We thank Sharon Rolando at the APHL for assistance with administration of the contract. We also thank Balasubra Swaminathan and Patti Fields at the Centers for Disease Control and Prevention for helpful scientific discussions and input on project direction.
Footnotes
Published ahead of print on 6 December 2006.
REFERENCES
- 1.Allix, C., P. Supply, and M. Fauville-Dufaux. 2004. Utility of fast mycobacterial interspersed repetitive unit-variable number tandem repeat genotyping in clinical mycobacteriological analysis. Clin. Infect. Dis. 39:783-789. [DOI] [PubMed] [Google Scholar]
- 2.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besser, J., J. Beebe, and B. Swaminathan. 2003. Investigation of foodborne and waterborne disease outbreaks, p. 162-181. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology. ASM Press, Washington, DC.
- 4.Boonmar, S., A. Bangtrakulnonth, S. Pornrunangwong, J. Terajima, H. Watanabe, K. Kaneko, and M. Ogawa. 1998. Epidemiological analysis of Salmonella enteritidis isolates from humans and broiler chickens in Thailand by phage typing and pulsed-field gel electrophoresis. J. Clin. Microbiol. 36:971-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2004. Outbreak of Salmonella serotype Enteritidis infections associated with raw almonds—United States and Canada, 2003-2004. Morb. Mortal. Wkly. Rep. 53:484-487. [PubMed] [Google Scholar]
- 6.Clark, C. G., T. M. A. C. Kruk, L. Bryden, Y. Hirvi, R. Ahmed, and F. G. Rodgers. 2003. Subtyping of Salmonella enterica serotype Enteritidis strains by manual and automated PstI-SphI ribotyping. J. Clin. Microbiol. 41:27-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2006. Molecular methods for bacterial strain typing; proposed guideline. Clinical and Laboratory Standards Institute document MM11-P. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Desai, M., E. J. Threlfall, and J. Stanley. 2001. Fluorescent amplified-fragment length polymorphism subtyping of the Salmonella enterica serovar Enteritidis phage type 4 clone complex. J. Clin. Microbiol. 39:201-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farlow, J., K. L. Smith, J. Wong, M. Abrams, M. Lytle, and P. Keim. 2001. Francisella tularensis strain typing using multiple-locus, variable-number tandem repeat analysis. J. Clin. Microbiol. 39:3186-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandes, S. A., A. C. Ghilardi, A. T. Tavechio, A. M. Machado, and A. C. Pignatari. 2003. Phenotypic and molecular characterization of Salmonella Enteritidis strains isolated in Sao Paulo, Brazil. Rev. Inst. Med. Trop. Sao Paulo 45:59-63. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez, J., A. Fica, G. Ebensperger, H. Calfullan, S. Prat, A. Fernandez, M. Alexandre, and I. Heitmann. 2003. Analysis of molecular epidemiology of Chilean Salmonella enterica serotype Enteritidis isolates by pulsed-field gel electrophoresis and bacteriophage typing. J. Clin. Microbiol. 41:1617-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hickman-Brenner, F. W., A. D. Stubbs, and J. J. Farmer III. 1991. Phage typing of Salmonella enteritidis in the United States. J. Clin. Microbiol. 29:2817-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilton, A. C., J. G. Banks, and C. W. Penn. 1996. Random amplification of polymorphic DNA (RAPD) of Salmonella: strain differentiation and characterization of amplified sequences. J. Appl. Bacteriol. 81:575-584. [DOI] [PubMed] [Google Scholar]
- 15.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isaacs, S., P. Sockett, J. Wilson, S. Styliadis, and A. Borczyk. 1997. Salmonella enteritidis phage type 4 in Ontario. Can. Commun. Dis. Rep. 23:177-181. [PubMed] [Google Scholar]
- 17.Johansson, A., J. Farlow, P. Larsson, M. Dukerich, E. Chambers, M. Bystrom, J. Fox, M. Chu, M. Forsman, A. Sjostedt, and P. Keim. 2004. Worldwide genetic relationships among Francisella tularensis isolates determined by multiple-locus variable-number tandem repeat analysis. J. Bacteriol. 186:5808-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klevytska, A. M., L. B. Price, J. M. Schupp, P. L. Worsham, J. Wong, and P. Keim. 2001. Identification and characterization of variable-number tandem repeats in the Yersinia pestis genome. J. Clin. Microbiol. 39:3179-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liebana, E., C. Clouting, L. Garcia-Migura, F. A. Clifton-Hadley, E. Lindsay, E. J. Threlfall, and R. H. Davies. 2004. Multiple genetic typing of Salmonella Enteritidis phage-types 4, 6, 7, 8 and 13a isolates from animals and humans in the UK. Vet. Microbiol. 100:189-195. [DOI] [PubMed] [Google Scholar]
- 21.Lin, A. W., M. A. Usera, T. J. Barrett, and R. A. Goldsby. 1996. Application of random amplified polymorphic DNA analysis to differentiate strains of Salmonella enteritidis. J. Clin. Microbiol. 34:870-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindstedt, B. A., E. Heir, E. Gjernes, and G. Kapperud. 2003. DNA fingerprinting of Salmonella enterica subsp. enterica serovar Typhimurium with emphasis on phage type DT104 based on variable number of tandem repeat loci. J. Clin. Microbiol. 41:1469-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindstedt, B. A., E. Heir, E. Gjernes, T. Vardund, and G. Kapperud. 2003. DNA fingerprinting of Shiga-toxin producing Escherichia coli O157 based on multiple-locus variable-number tandem-repeats analysis (MLVA). Ann. Clin. Microbiol. Antimicrob. 2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindstedt, B. A., T. Vardund, L. Aas, and G. Kapperud. 2004. Multiple-locus variable-number tandem-repeats analysis of Salmonella enterica subsp. enterica serovar Typhimurium using PCR multiplexing and multicolor capillary electrophoresis. J. Microbiol. Methods 59:163-172. [DOI] [PubMed] [Google Scholar]
- 25.Ling, M. L., and G. C. Wang. 2001. Epidemiological analysis of Salmonella enteritidis isolates in Singapore. J. Infect. 43:169-172. [DOI] [PubMed] [Google Scholar]
- 26.Liu, Y., M. A. Lee, E. E. Ooi, Y. Mavis, A. L. Tan, and H. H. Quek. 2003. Molecular typing of Salmonella enterica serovar Typhi isolates from various countries in Asia by a multiplex PCR assay on variable-number tandem repeats. J. Clin. Microbiol. 41:4388-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopes, V. C., B. T. Velayudhan, D. A. Halvorson, D. C. Lauer, R. K. Gast, and K. V. Nagaraja. 2004. Comparison of methods for differentiation of Salmonella enterica serovar Enteritidis phage type 4 isolates. Am. J. Vet. Res. 65:538-543. [DOI] [PubMed] [Google Scholar]
- 28.Magurran, A. 1988. Ecological diversity and its measurement, p. 8-45. Princeton University Press, Princeton, NJ.
- 29.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Medus, C., D. Boxrud, S. Abbott, R. Marcus, M. Park, T. McGivern, and D. Swerdlow. 2000. Abstr. Int. Conf. Emerg. Infect. Dis., abstr. 19/58.
- 30.Nei, M. 1987. Molecular evolutionary genetics. Columbia University Press, New York, NY.
- 31.Noller, A. C., M. C. McEllistrem, A. G. Pacheco, D. J. Boxrud, and L. H. Harrison. 2003. Multilocus variable-number tandem repeat analysis distinguishes outbreak and sporadic Escherichia coli O157:H7 isolates. J. Clin. Microbiol. 41:5389-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nygard, K., B. de Jong, P. J. Guerin, Y. Andersson, A. Olsson, and J. Giesecke. 2004. Emergence of new Salmonella Enteritidis phage types in Europe? Surveillance of infections in returning travellers. BMC Med. 2:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onteniente, L., S. Brisse, P. T. Tassios, and G. Vergnaud. 2003. Evaluation of the polymorphisms associated with tandem repeats for Pseudomonas aeruginosa strain typing. J. Clin. Microbiol. 41:4991-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patrick, M. E., P. M. Adcock, T. M. Gomez, S. F. Altekruse, B. H. Holland, R. V. Tauxe, and D. L. Swerdlow. 2004. Salmonella enteritidis infections, United States, 1985-1999. Emerg. Infect. Dis. 10:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramisse, V., P. Houssu, E. Hernandez, F. Denoeud, V. Hilaire, O. Lisanti, F. Ramisse, J. D. Cavallo, and G. Vergnaud. 2004. Variable number of tandem repeats in Salmonella enterica subsp. enterica for typing purposes. J. Clin. Microbiol. 42:5722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renders, N., L. Licciardello, C. Ijsseldijk, M. Sijmons, L. van Alphen, H. Verbrugh, and A. van Belkum. 1999. Variable numbers of tandem repeat loci in genetically homogeneous Haemophilus influenzae strains alter during persistent colonisation of cystic fibrosis patients. FEMS Microbiol. Lett. 173:95-102. [DOI] [PubMed] [Google Scholar]
- 37.Ribot, E. M., R. K. Wierzba, F. J. Angulo, and T. J. Barrett. 2002. Salmonella enterica serotype Typhimurium DT104 isolated from humans, United States, 1985, 1990, and 1995. Emerg. Infect. Dis. 8:387-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ridley, A. M., E. J. Threlfall, and B. Rowe. 1998. Genotypic characterization of Salmonella enteritidis phage types by plasmid analysis, ribotyping, and pulsed-field gel electrophoresis. J. Clin. Microbiol. 36:2314-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodrigue, D. C., D. N. Cameron, N. D. Puhr, F. W. Brenner, M. E. St. Louis, I. K. Wachsmuth, and R. V. Tauxe. 1992. Comparison of plasmid profiles, phage types, and antimicrobial resistance patterns of Salmonella enteritidis isolates in the United States. J. Clin. Microbiol. 30:854-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schouls, L. M., H. G. van der Heide, L. Vauterin, P. Vauterin, and F. R. Mooi. 2004. Multiple-locus variable-number tandem repeat analysis of Dutch Bordetella pertussis strains reveals rapid genetic changes with clonal expansion during the late 1990s. J. Bacteriol. 186:5496-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schouls, L. M., A. van der Ende, I. van de Pol, C. Schot, L. Spanjaard, P. Vauterin, D. Wilderbeek, and S. Witteveen. 2005. Increase in genetic diversity of Haemophilus influenzae serotype b (Hib) strains after introduction of Hib vaccination in The Netherlands. J. Clin. Microbiol. 43:2741-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scott, F., J. Threlfall, J. Stanley, and C. Arnold. 2001. Fluorescent amplified fragment length polymorphism genotyping of Salmonella Enteritidis: a method suitable for rapid outbreak recognition. Clin. Microbiol. Infect. 7:479-485. [DOI] [PubMed] [Google Scholar]
- 43.Shannon, C. E., and W. Weaver. 1949. The mathematical theory of communication. University of Illinois Press, Urbana.
- 44.Spurgiesz, R. S., T. N. Quitugua, K. L. Smith, J. Schupp, E. G. Palmer, R. A. Cox, and P. Keim. 2003. Molecular typing of Mycobacterium tuberculosis by using nine novel variable-number tandem repeats across the Beijing family and low-copy-number IS6110 isolates. J. Clin. Microbiol. 41:4224-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Struelens, M. J. 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2-11. [DOI] [PubMed] [Google Scholar]
- 46.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Titze-de-Almeida, R., R. J. Willems, J. Top, I. P. Rodrigues, R. F. Ferreira, H. Boelens, M. C. Brandileone, R. C. Zanella, M. S. Felipe, and A. van Belkum. 2004. Multilocus variable-number tandem-repeat polymorphism among Brazilian Enterococcus faecalis strains. J. Clin. Microbiol. 42:4879-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Belkum, A., S. Scherer, W. van Leeuwen, D. Willemse, L. van Alphen, and H. Verbrugh. 1997. Variable number of tandem repeats in clinical strains of Haemophilus influenzae. Infect. Immun. 65:5017-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wachsmuth, I. K., J. A. Kiehlbauch, C. A. Bopp, D. N. Cameron, N. A. Strockbine, J. G. Wells, and P. A. Blake. 1991. The use of plasmid profiles and nucleic acid probes in epidemiologic investigations of foodborne, diarrheal diseases. Int. J. Food Microbiol. 12:77-89. [DOI] [PubMed] [Google Scholar]
- 50.Ward, L. R., J. D. de Sa, and B. Rowe. 1987. A phage-typing scheme for Salmonella enteritidis. Epidemiol. Infect. 99:291-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yazdankhah, S. P., B. A. Lindstedt, and D. A. Caugant. 2005. Use of variable-number tandem repeats to examine genetic diversity of Neisseria meningitidis. J. Clin. Microbiol. 43:1699-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zar, J. H. 1984. Biostatistical analysis, p. 32-36. Prentice-Hall, Englewood Cliffs, NJ.