Abstract
An optimized set of 24 mycobacterial interspersed repetitive-unit-variable-number tandem-repeat (MIRU-VNTR) loci, including a discriminatory subset of 15 loci, has recently been defined for the typing of Mycobacterium tuberculosis. Here, we evaluated the performances of this MIRU-VNTR typing system in combination with spoligotyping for the detection of transmission chains in a population-based study comprising 91% of culture-confirmed tuberculosis patients reported in 2003 in Hamburg, Germany. Of the 154 isolates investigated, more than 90% had high IS6110 copy numbers (≥6). IS6110 restriction fragment length polymorphism (RFLP) typing resulted in 13 clusters, 5 of which had a confirmed epidemiological link. All five, as well as six of the eight IS6110 clusters with no identified epidemiological link, were perfectly matched by MIRU-VNTR typing with the 24 loci. Two IS6110 clusters were split by differences into 6 to 12 MIRU-VNTR loci, clearly supporting the absence of a link, as judged by contact tracing data. In contrast, only one MIRU-VNTR cluster, grouping what were probably epidemiologically unlinked isolates, was split by IS6110 RFLP. However, these isolates were also distinguished by spoligotyping. Both the optimized 24-locus and 15-locus sets thus showed a comparable to slightly better predictive value, especially when combined with spoligotyping, than the current gold standard IS6110 RFLP for the study of tuberculosis transmission in Hamburg. Because the epidemiological characteristics of this setting are similar to those of many developed countries, these results support the wide applicability of this real-time genotyping approach for population-based studies of M. tuberculosis transmission.
Population-based molecular epidemiological studies of tuberculosis (TB) aim to identify pathways and predictors of ongoing TB transmission by determining the proportion of patients with identical isolates (clustering rate), as defined by molecular typing at regional or national scales. IS6110 restriction fragment length polymorphism (RFLP) typing of Mycobacterium tuberculosis is the gold standard method for this purpose (30). Consistently, M. tuberculosis outbreak isolates often show identical IS6110 RFLP patterns, and clustering by identical patterns is associated with well-identified risk factors for transmission, whereas unrelated patients often have isolates with different IS6110 fingerprints (see, for example, references 6, 21, 31, and 33).
However, IS6110 fingerprinting is laborious and requires weeks of M. tuberculosis culturing, which limits the possibilities to use typing prospectively for more efficient TB control. Furthermore, comparison of the fingerprints from large data sets requires highly standardized experimental and computerized procedures (30). These problems complicate the determination of clustering rates in population-based studies and the exchange of data.
Typing based on PCR amplification of multiple loci containing mycobacterial interspersed repetitive-unit-variable-number tandem repeats (MIRU-VNTR) offers a potential solution to these drawbacks. This method is highly reproducible and yields portable results. It is much faster than IS6110-RFLP typing, since it can be directly applied to crude extracts from colonies or early culture pellets. Among the different described sets of MIRU-VNTR loci (8, 10, 14, 17, 20, 22, 27), a system based on 12 loci (15, 25, 27), is now the most widely used in clinical mycobacteriology (see, for example, reference 1) and for local outbreak investigation (11). Genotypes based on these 12 loci are highly stable among epidemiologically linked isolates but sufficiently diverse to generate a resolution approaching that of IS6110-RFLP in some sets of isolates from low-incidence settings or from diverse geographic origins (2, 15, 25).
A population-based study indicated that the use of this 12-locus-based MIRU-VNTR typing combined with spoligotyping as a first-line approach provides adequate discrimination in most cases for large-scale genotyping of M. tuberculosis in the United States (4). Consistent with a generally lower rate of change of these 12-locus patterns than that of IS6110-RFLP (15, 18, 25), secondary IS6110 fingerprinting of the isolates clustered by this combined strategy is nevertheless necessary to reach the maximal resolution needed in some situations (4, 18). However, another investigation suggested that this MIRU-VNTR typing combined with spoligotyping was inadequate for population-based studies because it provided a too-low resolution power and, more surprisingly, because MIRU-VNTR genotypes were apparently too frequently unstable among presumably related isolates (19). However, these parameters were determined by taking clustering defined by IS6110-RFLP as a sole reference, in the absence of any contact tracing data. Since it is known that IS6110-RFLP does not per se unambiguously define all related or unrelated isolates (see, for example, references 29, 32, and 34), unequivocal interpretation of all of the divergences between the typing methods in that study (19) was not possible.
In a recent study based on an worldwide collection of tubercle bacillus isolates (24), we defined an optimized set of 24 MIRU-VNTR loci, including a subset of 15 discriminatory loci proposed to be used as a first-line typing method. These 24- and 15-locus sets reliably improved the discrimination of M. tuberculosis isolates compared to the original 12-locus set. Here, we evaluated the performances of this optimized set in combination with spoligotyping versus IS6110-RFLP for analysis of clustering among isolates collected from an ongoing population-based study in Hamburg, Germany. The clustering results generated by all methods were compared to individual contact tracing data.
MATERIALS AND METHODS
Study population and data collection.
The study includes patients with culture-confirmed TB that were reported to any of the seven district public health departments of Hamburg between 1 January and 31 December 2003. Hamburg is one of the German federal states and, with 1.7 million residents, it is the second largest city of Germany. Case data were collected prospectively by trained public health staff by using a standardized questionnaire, as described previously (6, 7). Briefly, information was obtained on gender, date and country of birth, nationality, immigration status (if relevant), number of years of residency in Hamburg or elsewhere in Germany, present address (or whether the patient was homeless), whether the patient was living in a health care institution or any public institution, the nature of the patient's employment, socioeconomic status, any previous known exposure to other persons with TB (especially within the 6 months before development of any symptoms), and the identity of the patient's household contacts and respective close contacts in occupational or crowded settings, if relevant. In addition, a thorough reinvestigation of clustered cases was conducted that included retrospective patient interviews when possible.
Bacterial strains and drug susceptibility testing.
Of the 169 patients with culture-confirmed TB notified in the study period, mycobacterial strains from 154 patients (91%) were available for the analysis. Primary isolation and culturing of mycobacterial isolates were performed as described previously (6). All isolates were identified as M. tuberculosis complex by using the Genotype MTBC kit as instructed by the manufacturer (HAIN Lifescience, GmbH, Nehren, Germany). Drug susceptibility was determined with the BACTEC MGIT 960TB according to the instructions of the supplier (Becton Dickinson Microbiology Systems, Cockeysville, MD).
IS6110-RFLP and spoligotyping.
Extraction of DNA from mycobacterial strains and DNA fingerprinting using IS6110 as a probe was performed according to a internationally standardized protocol as described elsewhere (16, 30). PvuII-digested total DNA of reference strain Mt.14323 was included in each Southern blot experiment as an external size standard and was used for quality control of IS6110 RFLP experiments. IS6110 RFLP patterns were analyzed by using Bionumerics (Applied Maths, St-Martin-Latem, Belgium) as described previously (16, 30). Spoligotyping of isolates was performed as described by Kamerbeek et al. (12).
MIRU-VNTR genotyping.
MIRU-VNTR genotyping was performed blindly, before decoding of IS6110 RFLP and spoligotyping results. Twenty-four loci were amplified by PCR as described elsewhere (14, 17, 20, 23, 24, 27). Briefly, analyses were performed by using multiplex PCR, Rox-labeled MapMarker 1000 size standard (BioVentures, Murfreesboro, TN) and ABI 3100 and ABI 3730-XL sequencers. Sizing of the PCR fragments and assignment of the various VNTR alleles were done by using customized GeneScan and Genotyper or Genemapper software packages (PE Applied Biosystems).
PCR mixtures were prepared as follows, using 96-well plates and a HotStartTaq DNA polymerase kit (QIAGEN, Hilden, Germany). DNA (2 ng) was added to a final volume of 20 μl containing 0.08 μl of DNA polymerase (0.4 U); 4 μl of Q-solution; 0.2 mM concentrations of dATP, dCTP, dGTP, and dTTP (Pharmacia, Uppsala, Sweden); 2 μl of PCR buffer; 1.5 to 3.0 mM MgCl2; 0.4 μM concentrations of each unlabeled oligonucleotide; and 0.04 to 0.4 μM concentrations of labeled oligonucleotides. Negative controls consisted of the PCR performed on reaction mixtures lacking mycobacterial DNA.
The thermocycler programs were identical for the eight multiplex reactions. The PCRs were carried out by using a Hybaid PCR Express cycler (Hybaid, Ashford, Great Britain) or PTC 200 cycler (MJ Research, Watertown, MA) starting with a denaturing step of 15 min at 95°C, followed by 40 cycles of 1 min at 94°C, 1 min at 59°C, and 1 min 30 s at 72°C. The reactions were terminated by incubating 10 min at 72°C.
Clustering analysis.
Clustering by the different typing methods was analyzed by using Bionumerics (Applied Maths). IS6110 RFLP clusters were defined based on fully identical patterns (the same number of IS6110 bands at identical positions [position tolerance, 1.2%]). IS6110 RFLP and spoligotyping and MIRU-VNTR dendrograms were generated by using the unweighted pair group method with arithmetic averages and the Dice or the categorical coefficient (IS6110 RFLP) (spoligotyping and MIRU-VNTR). The clustering rate was defined as (nc − c)/n, where nc is the total number of clustered cases, c is the number of clusters, and n is the total number of cases in the sample (21).
RESULTS
Study population.
Between 1 January and 31 December 2003, 216 TB patients were reported to any of the seven district public health departments of Hamburg. Of these, 169 had a culture-confirmed TB. A total of 154 of the corresponding isolates were available for genotyping and were included in this investigation. In the study population, the majority of patients were male (n = 99, 64%), 68 patients (44%) were born in Germany, and 10 showed resistance to antitubercular drugs (6.5%); however, only two cases (1.3%) were reported with resistance to at least isoniazid and rifampin and thus were multidrug resistant.
Genotype distribution.
Among the 154 isolates analyzed, 90.9% (n = 140) had IS6110 RFLP profiles with high copy numbers (≥6 IS6110 bands), whereas the remaining 9.1% (n = 14) had fingerprints with one to five bands (see Fig. S1 in the supplemental material). The spoligotyping and MIRU-VNTR typing results congruently indicated a marked prevalence of Haarlem genotypes (37%, n = 55) in the collection, followed by CAS/Delhi, East-African Indian, Beijing, and LAM genotypes (from 7.8 to 5.8% or 12 to 9 isolates for each lineage). The remaining isolates displayed various spoligo-prototypes assigned to genetically less-consistent or not well-defined clades such as T (n = 22) and S (n = 2) or to unassigned genotypes (n = 33).
Discriminatory power and clustering rates.
Based on IS6110 fingerprinting a total of 39 isolates were grouped within 13 clusters (see Fig. S1a in the supplemental material), thus generating 115 unique and 39 shared IS6110 DNA fingerprints. By applying MIRU-VNTR typing with the full set of 24 loci (Table 1) , a slightly better resolution with 117 unique and 37 clustered isolates (11 clusters) was observed (see Fig. S1b in the supplemental material). However, both methods yielded identical clustering rates of 16.9% due to minor differences in cluster compositions (see below and Table 2). The use of the discriminatory subset of 15 MIRU-VNTR loci (Table 1) only marginally affected the resolution, with a clustering rate of 17.5% that resulted from the addition of a single cluster containing only two isolates (indicated by arrows in Fig. S1a and b in the supplemental material).
TABLE 1.
Composition of the locus sets
| Set and multiplex | Locusa | Aliasb |
|---|---|---|
| 15 Locus (discriminatory) | ||
| Mix 1 | 580 | MIRU 4, ETR D |
| 2996 | MIRU 26 | |
| 802 | MIRU 40 | |
| Mix 2 | 960 | MIRU 10 |
| 1644 | MIRU 16 | |
| 3192 | MIRU 31, ETR E | |
| Mix 3 | 424 | Mtub04 |
| 577 | ETR C | |
| 2165 | ETR A | |
| Mix 4 | 2401 | Mtub30 |
| 3690 | Mtub39 | |
| 4156 | QUB-4156 | |
| Mix 5 | 2163b | QUB-11b |
| 1955 | Mtub21 | |
| 4052 | QUB-26 | |
| 24 Locus (full set) | ||
| Mix 6 | 154 | MIRU 2 |
| 2531 | MIRU 23 | |
| 4348 | MIRU 39 | |
| Mix 7 | 2059 | MIRU 20 |
| 2687 | MIRU 24 | |
| 3007 | MIRU 27, QUB-5 | |
| Mix 8 | 2347 | Mtub29 |
| 2461 | ETR B | |
| 3171 | Mtub34 |
Corresponds to positions on the H37Rv chromosome in kbp.
ETR, exact tandem repeat, QUB, Queen's University of Belfast.
TABLE 2.
Clustering results by genotyping methods
| Methoda | No. of unique isolates | No. of clustered isolates | No. of clusters | No. of distinct types | Clustering rate (%)b |
|---|---|---|---|---|---|
| Spoligotyping | 56 | 98 | 16 | 72 | 53.2 |
| MIRU-VNTR 12 old | 84 | 70 | 20 | 104 | 32.5 |
| MIRU-VNTR 12 old + 3 ETR | 103 | 51 | 13 | 116 | 24.7 |
| MIRU-VNTR 12 old + spoligotyping | 106 | 48 | 14 | 120 | 22.1 |
| IS6110-RFLP | 115 | 39 | 13 | 128 | 16.9 |
| Low-copy-no. isolates | 8 | 6 | 2 | 10 | NR |
| High-copy-no. isolates | 107 | 33 | 11 | 118 | NR |
| MIRU-VNTR 15 | 115 | 39 | 12 | 127 | 17.5 |
| MIRU-VNTR 24 | 117 | 37 | 11 | 128 | 16.9 |
| MIRU-VNTR 15 + spoligotyping | 120 | 34 | 11 | 131 | 14.9 |
| MIRU-VNTR 24 + spoligotyping | 120 | 34 | 11 | 131 | 14.9 |
MIRU-VNTR 12 old, original set of 12 MIRU-VNTR loci as described previously (26, 27), MIRU-VNTR 12 old + 3 ETR, original set of 12 MIRU-VNTR loci plus three nonredundant exact-tandem-repeat loci (8), MIRU-VNTR 15 and 24, discriminatory subset of 15 loci and full set of 24 loci (Table 1) (24).
NR, not relevant.
Spoligotyping slightly increased the resolution of MIRU-VNTR typing based on the 24 loci by discriminating three isolates clustered by this MIRU-VNTR set. These three isolates had distinct IS6110 RFLP profiles (see MIRU cluster 9 and 9b in Fig. S1b in the supplemental material and Fig. 2 [see below]). The two additional isolates clustered by the 15 MIRU-VNTR loci (see above) were also discriminated by spoligotyping (see arrows in Fig. S1a and b in the supplemental material). Therefore, the maximal resolution was achieved by combining MIRU-VNTR typing based either on 15 or 24 loci with spoligotyping (clustering rate of 14.9%).
FIG. 2.
MIRU-VNTR SLVs. Isolates differing by a single MIRU-VNTR locus (boxed) out of the 24 loci are shown, along with the corresponding IS6110 RFLP and spoligotyping profiles. Isolate 10041/03 displayed SLVs (shown by a single box or two separate connected boxes) with three different isolates. Similarly, the isolates in MIRU-VNTR cluster 9+9b (see the text) displayed SLVs (shown by a single box or two separate connected boxes) with two different isolates.
In comparison, the use of MIRU-VNTR typing based either on the 12 original MIRU-VNTR loci alone, in combination with three nonredundant exact tandem repeat (ETR) loci (8) or with spoligotyping, resulted in a significant increase of number of clusters and clustered isolates (with clustering rates of 32.5, 24.7, or 22.1%, respectively; Table 2). This increase was principally due to lower discrimination of the 12 MIRU-VNTR loci among the 55 isolates of the predominant Haarlem lineage. Of these, 39 (71%) were clustered by 12 loci MIRU-VNTR typing, whereas 24 (44%) or 22 (40%) isolates were clustered by IS6110 DNA fingerprinting or by MIRU-VNTR based on 24 loci, respectively (data not shown).
Congruence of cluster analysis and epidemiological links.
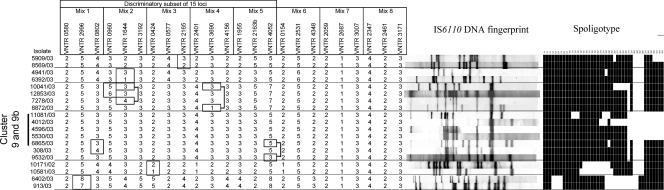
Of the 13 IS6110 RFLP clusters grouping a total of 39 isolates, 10 including 32 isolates were found to be completely identical by the set of 24 MIRU-VNTR loci (Fig. 1 and Fig. S1 in supplemental material). These 10 clusters corresponded to five Haarlem, one Beijing, one Latin American Mediterranean, one Central Asian/Delhi, and two unassigned genotypes. In five cases, epidemiological links among clustered patients were confirmed by contact tracing.
FIG. 1.
IS6110 DNA fingerprint patterns, spoligotype patterns, and MIRU-VNTR copy numbers of the 39 strains grouped in 13 IS6110 clusters. In addition, the genotypes of the three strains additionally clustered by identical MIRU patterns but differentiated by spoligotyping (cluster 9b) are shown. Isolates of IS6110 clusters 1 and 12 without epidemiological link (see the text) are clearly distinguished by differences in their MIRU-VNTR profiles (boxed).
Of the three remaining IS6110 RFLP clusters, one grouping two isolates with a Haarlem genotype showed a partial overlap with a cluster defined by the 24 MIRU-VNTR loci (cluster 9 and 9b; Fig. 1). Three other isolates were additionally clustered with the former isolates by these markers. These isolates showed clear differences of at least three IS6110 bands in their RFLP profiles either mutually or with the two isolates clustered by IS6110 RFLP. These three isolates were also distinguished from one another and from the two clustered isolates by spoligotyping. No evidence of epidemiological links could be found among the patients in this cluster.
Conversely, three isolates with an East African Indian genotype and grouped within a cluster by their single IS6110 band fingerprint (cluster 1 in Fig. 1) were distinguished by mutual MIRU-VNTR typing, with differences from 9 to 12 loci of the 24. This result was confirmed by spoligotypes mutually distinct by two blocks of spacers and compatible with the absence of evidence for epidemiological link among the respective patients.
Finally, cluster 12, grouping two isolates with an identical seven-band IS6110 fingerprint and an identical Haarlem spoligotype, was split by seven different MIRU-VNTR loci of the 24. Patient 1 of this cluster was an 31-year-old Algerian guest-worker without any history of previous TB disease and living in Germany since 1989. Patient 2 was a 46-year-old German whose source of infection could be traced to his brother suffering from TB in 1997. Although intensive prospective and retrospective interviews have been performed, no epidemiological connection between both patients nor with the brother of patient 2 could be established. It is therefore likely that patient 1 acquired his infection with this prevalent Haarlem genotype strain during his long stay in Germany independently from the other patients and that the development of disease during the study period is only coincidental.
Epidemiological significance of MIRU-VNTR SLV.
In a companion study (24), the subset of 15 discriminatory loci was designed to concentrate the markers with the most frequent involvement in single-, double-, or triple-locus variations among closely related isolates from different lineages from cosmopolitan origins. Therefore, we examined the distribution of such events, limited to single-locus variations (SLVs) in this case, among the full set of 24 loci or within the 15-locus discriminatory subset in the Hamburg population. We then examined their significance by comparing them by IS6110 RFLP analysis.
When the 24 loci were considered, only nine cases of SLVs were detected among the 154 isolates (Fig. 2). These SLVs all involved markers also present in the 15-locus subset, i.e., twice locus MIRU 16 and once MIRU 10, 26, 40, and VNTR 0424, ETR A (2165), QUB-26 (4052), and VNTR 3690. These SLVs were found five times between Haarlem isolates, once between CAS/Delhi isolates, and three times between isolates with a T or an unassigned spoligotype. Consistent with the results of cluster analysis (see above), all of the isolates differing by a SLV were also defined as unique by IS6110 RFLP.
When only the 15 loci composing the discriminatory subset were considered, six additional SLVs were detected, involving locus MIRU 10 twice and MIRU 26, MIRU 40, QUB 2163b, and QUB 26 (4052) once. Five of these SLVs were found among Beijing isolates, and one was between a T5 and an unassigned genotype. Again, none of these respective isolates were found within IS6110 RFLP clusters (data not shown).
DISCUSSION
In this model population representing 91% of culture-positive TB cases collected in 2003 in Hamburg, the optimized MIRU-VNTR set showed a comparable to slightly better predictive value for the study of TB transmission than the current gold standard IS6110 RFLP, especially when combined with spoligotyping. The conditions in which this comparison was performed were stringent, since the isolate population displayed more than 90% of IS6110 high-copy-number fingerprints, for which IS6110 RFLP is especially discriminatory, and 37% of isolates from the single Haarlem lineage for which the original set of 12 MIRU-VNTR loci showed a relatively low discriminatory power.
Isolates clustered with identical IS6110 fingerprints are generally assumed to be clonal if they contain high copy numbers of IS6110 (≥6) and, depending on the context, considered to be prognostic of ongoing transmission events. In the present study, IS6110 RFLP typing resulted in 13 clusters, 5 of which had a confirmed epidemiological link. These five IS6110 clusters were perfectly matched by MIRU-VNTR typing over the 24 loci used. Moreover, six of the eight IS6110 clusters with no identified epidemiological link were also identically retrieved with MIRU-VNTR typing. In these cases, the perfect concordance between two highly discriminatory molecular methods does suggest that the patients involved were infected by the same strain, despite the absence of confirmation by contact tracing. This lack of confirmation can then be explained by the occurrence of transmission through casual contacts, barhopping, or illegal activities (which are difficult to detect by traditional investigation) (5, 28, 33), transmission chains not covered by the study period, or reactivation of a previous infection with an endemic strain (3, 9).
These findings confirm previous reports on the high stability of MIRU-VNTR types (based on 12, 15, or 24 loci) among epidemiologically linked isolates and in IS6110 clusters (4, 11, 13, 15, 18, 24, 29). They are in contrast to a single study, which reported changes in MIRU-VNTR patterns among 48% of isolates from IS6110 clusters (in addition to spoligotype changes among 17% of isolates from IS6110 clusters) (19). The significance of these latter results remains unclear in the absence of contact tracing data.
Actually, several other studies, integrating contact tracing data, showed that not all clusters based on IS6110 RFLP patterns, even when multibanded, necessarily reflect clonal transmission of TB (3, 4, 13, 28, 29). Systematic assessment of the epidemiological significance of IS6110-RFLP clusters revealed a proportion of 14% of false IS6110 clusters in a Dutch population-based context (29). These clusters without any evidence of epidemiological link after thorough epidemiological investigation differed by two to six loci from the 15-locus discriminatory subset (24, 29). Here, in the Hamburg population, one cluster of two isolates with an identical IS6110 high-copy-number fingerprint and an identical spoligotype was split by as many as seven MIRU-VNTR loci, four of which are part of the 15-locus subset. The IS6110 RFLP, spoligotyping, and MIRU-VNTR results were repeated using the same original DNA samples, ruling out trivial laboratory errors. Both prospective and retrospective contact tracing provided no evidence of any epidemiological link among the patients involved (see Results). Therefore, the differences in four of the 15-locus subset observed here, reinforced by three additional differences among auxiliary loci with a lower evolutionary rate, leave little doubt that the patients were falsely clustered by IS6110 high-copy-number fingerprints. More trivially, a second IS6110 RFLP cluster, including three isolates with a single IS6110 band, was split by 6 to 12 differences in the 15- or the 24-locus set, as well as by spoligotyping. These two examples show how the interrogation of 15 or even 24 independent markers can increase the degree of confidence for reliably excluding recent transmission, especially if the results obtained with IS6110 RFLP typing are conflicting.
Even differences by a single MIRU-VNTR locus remain strongly predictive of the absence of a link, in contrast to results reported by Scott et al. (19). In the present study, a total of 15 SLV events were detected within the 15-locus subset. None of the corresponding isolates distinguished by these SLVs was clustered by IS6110 RFLP, which corroborates the absence of a link. These results are fully consistent with our recent evaluation of MIRU-VNTR stability using large sets of epidemiologically linked or clonal isolates (24). We estimated the risk of erroneously excluding from a cluster an isolate showing an SLV in the 15-locus subset to be only of 5 to 6%. This level is just below the statistically predicted threshold of detection in the present study (6.7%, corresponding to 1 epidemiologically erroneous SLV among a total of 15 SLV events).
In all, only three isolates among the 115 defined as unique by IS6110 RFLP were clustered by the 24 MIRU-VNTR loci (see cluster 9b in Fig. 2). These three isolates shared common IS6110 banding patterns typical of the Haarlem lineage, demonstrating their close genetic relationships. However, these three isolates judged unique by IS6110 RFLP were also distinguished by spoligotyping, clearly indicating the absence of a link in agreement with contact tracing. Moreover, the use of the 15-locus subset resulted in the addition of a single cluster containing only two other isolates, thus only marginally affecting the resolution power obtained with the full set of 24 loci. Interestingly, these two isolates were also discriminated by spoligotyping (see arrows in Fig. S1a in the supplemental material). These observations support the design of the 15-locus subset, which concentrates most of the resolution obtained with the full 24 loci across different M. tuberculosis lineages. Furthermore, these findings show that the combined use of spoligotyping can add marginal but still meaningful resolution to the optimized 15- or 24-MIRU-VNTR locus sets (24), resulting in the discrimination of all of the isolates already predicted as unique by IS6110 RFLP in the present case. In comparison, 9 or 14 isolates among the 115 or the 120 defined as unique by IS6110 RFLP or by combined optimized MIRU-VNTR and spoligotyping, respectively, were clustered by the use of the 12 original MIRU-VNTR loci with spoligotyping. This false clustering mainly resulted from lower discrimination among the predominant Haarlem isolates and would have led to an overestimate of the TB transmission rate in Hamburg, albeit to a lower degree than that reported for the use of these 12 original loci with spoligotyping in Montreal (19).
Thus, if the specificity and sensitivity for cluster analysis are calculated as described previously (19) (i.e., a proportion of cases classified as unique by one method among cases classified as unique by another and a proportion of cases identically clustered between two methods, respectively), but by additionally taking into account the contact tracing data, MIRU-VNTR typing based on the 15 or the 24 loci showed an overall specificity of 95.7 and 97.4% compared to IS6110 RFLP. The specificity went up to 100% when either of the two sets was combined with spoligotyping. In addition, both MIRU-VNTR typing sets showed a sensitivity of 100% to detect the proven or possibly epidemiologically linked IS6110 clusters, excluding the most likely false IS6110 clusters 1 and 12 (see above). Conversely, IS6110 RFLP also showed a sensitivity of 100% but a specificity of only 95.8% (due to false IS6110 clusters 1 and 12) compared to the combination of MIRU-VNTR and spoligotyping.
The study period for the comparison of these molecular methods as tools to study ongoing TB transmission was limited to a single year. The analysis of the ongoing transmission itself requires longer periods (usually 2 or 3 years). Over longer periods, it has been noticed that the rate of molecular clustering often increases because, among other reasons, transmission chains are more efficiently covered (9). However, because our results are based on an almost complete population representative of 1 year, we expect that this phenomenon will occur with both MIRU-VNTR and IS6110 RFLP fingerprinting, without much affecting the relative performances of the two methods.
In conclusion, our population-based investigation supports the wide applicability of a combination of the newly defined 15 loci MIRU-VNTR typing and spoligotyping as a real-time approach for population-based studies of TB transmission, at least in the numerous low-incidence settings with epidemiological characteristics similar to those of Hamburg. In areas with a higher incidence of TB, where the epidemiological interpretation of the results may be more difficult, this approach can be used at least as a reliable exclusion method (i.e., to identify most likely unrelated isolates by genotypic differences) to get an estimate of the maximal rates of ongoing TB transmission. The 15 or 24 MIRU-VNTR typing combined with spoligotyping represents the first PCR-based method with operating parameters (specificity and sensitivity) comparable to those of the gold-standard method IS6110 fingerprinting in a population-based investigation and thus can be used as a stand-alone approach to the study of TB transmission. Because of its speed and ease of data exchange and comparison, this method may actually become the new gold standard method.
Supplementary Material
Acknowledgments
We thank I. Radzio, T. Ubben, P. Vock (FZ Borstel), and E. Willery (Lille) for excellent technical assistance.
This study was supported in part by the Germany Ministry of Health, Berlin, Germany; the Robert Koch Institute, Berlin, Germany; INSERM and Institut Pasteur de Lille, France; and the European Community (grant QLK2-CT-2000-00630). M.C.O. held a fellowship from the CAPES, Brazil. P.S. is a Chercheur du Centre National de Recherche Scientifique.
Footnotes
Published ahead of print on 27 December 2006.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Allix, C., P. Supply, and M. Fauville-Dufaux. 2004. Utility of fast mycobacterial interspersed repetitive unit-variable number tandem repeat genotyping in clinical mycobacteriological analysis. Clin. Infect. Dis. 39:783-789. [DOI] [PubMed] [Google Scholar]
- 2.Blackwood, K. S., J. N. Wolfe, and A. M. Kabani. 2004. Application of mycobacterial interspersed repetitive unit typing to Manitoba tuberculosis cases: can restriction fragment length polymorphism be forgotten? J. Clin. Microbiol. 42:5001-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braden, C. R., G. L. Templeton, M. D. Cave, S. Valway, I. M. Onorato, K. G. Castro, D. Moers, Z. Yang, W. W. Stead, and J. H. Bates. 1997. Interpretation of restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates from a state with a large rural population. J. Infect. Dis. 175:1446-1452. [DOI] [PubMed] [Google Scholar]
- 4.Cowan, L. S., L. Diem, T. Monson, P. Wand, D. Temporado, T. V. Oemig, and J. T. Crawford. 2005. Evaluation of a two-step approach for large-scale, prospective genotyping of Mycobacterium tuberculosis isolates in the United States. J. Clin. Microbiol. 43:688-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diel, R., S. Rusch-Gerdes, and S. Niemann. 2004. Molecular epidemiology of tuberculosis among immigrants in Hamburg, Germany. J. Clin. Microbiol. 42:2952-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diel, R., S. Schneider, K. Meywald-Walter, C. M. Ruf, S. Rusch-Gerdes, and S. Niemann. 2002. Epidemiology of tuberculosis in Hamburg, Germany: long-term population-based analysis applying classical and molecular epidemiological techniques. J. Clin. Microbiol. 40:532-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diel, R., A. Seidler, A. Nienhaus, S. Rusch-Gerdes, and S. Niemann. 2005. Occupational risk of tuberculosis transmission in a low incidence area. Respir. Res. 6:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 9.Glynn, J. R., J. Bauer, A. S. de Boer, M. W. Borgdorff, P. E. Fine, P. Godfrey-Faussett, E. Vynnycky, et al. 1999. Interpreting DNA fingerprint clusters of Mycobacterium tuberculosis. Int. J. Tuberc. Lung. Dis. 3:1055-1060. [PubMed] [Google Scholar]
- 10.Goyal, M., D. Young, Y. Zhang, P. A. Jenkins, and R. J. Shaw. 1994. PCR amplification of variable sequence upstream of katG gene to subdivide strains of Mycobacterium tuberculosis complex. J. Clin. Microbiol. 32:3070-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawkey, P. M., E. G. Smith, J. T. Evans, P. Monk, G. Bryan, H. H. Mohamed, M. Bardhan, and R. N. Pugh. 2003. Mycobacterial interspersed repetitive unit typing of Mycobacterium tuberculosis compared to IS6110-based restriction fragment length polymorphism analysis for investigation of apparently clustered cases of tuberculosis. J. Clin. Microbiol. 41:3514-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwara, A., R. Schiro, L. S. Cowan, N. E. Hyslop, M. F. Wiser, S. Roahen Harrison, P. Kissinger, L. Diem, and J. T. Crawford. 2003. Evaluation of the epidemiologic utility of secondary typing methods for differentiation of Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 41:2683-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Fleche, P., M. Fabre, F. Denoeud, J. L. Koeck, and G. Vergnaud. 2002. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazars, E., S. Lesjean, A. L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niemann, S., S. Rusch-Gerdes, and E. Richter. 1997. IS6110 fingerprinting of drug-resistant Mycobacterium tuberculosis strains isolated in Germany during 1995. J. Clin. Microbiol. 35:3015-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roring, S., A. Scott, D. Brittain, I. Walker, G. Hewinson, S. Neill, and R. Skuce. 2002. Development of variable-number tandem repeat typing of Mycobacterium bovis: comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J. Clin. Microbiol. 40:2126-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savine, E., R. M. Warren, G. D. van der Spuy, N. Beyers, P. D. van Helden, C. Locht, and P. Supply. 2002. Stability of variable-number tandem repeats of mycobacterial interspersed repetitive units from 12 loci in serial isolates of Mycobacterium tuberculosis. J. Clin. Microbiol. 40:4561-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott, A. N., D. Menzies, T. N. Tannenbaum, L. Thibert, R. Kozak, L. Joseph, K. Schwartzman, and M. A. Behr. 2005. Sensitivities and specificities of spoligotyping and mycobacterial interspersed repetitive unit-variable-number tandem repeat typing methods for studying molecular epidemiology of tuberculosis. J. Clin. Microbiol. 43:89-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skuce, R. A., T. P. McCorry, J. F. McCarroll, S. M. Roring, A. N. Scott, D. Brittain, S. L. Hughes, R. G. Hewinson, and S. D. Neill. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148:519-528. [DOI] [PubMed] [Google Scholar]
- 21.Small, P. M., P. C. Hopewell, S. P. Singh, A. Paz, J. Parsonnet, D. C. Ruston, G. F. Schecter, C. L. Daley, and G. K. Schoolnik. 1994. The epidemiology of tuberculosis in San Francisco: a population-based study using conventional and molecular methods. N. Engl. J. Med. 330:1703-1709. [DOI] [PubMed] [Google Scholar]
- 22.Smittipat, N., P. Billamas, M. Palittapongarnpim, A. Thong-On, M. M. Temu, P. Thanakijcharoen, O. Karnkawinpong, and P. Palittapongarnpim. 2005. Polymorphism of variable-number tandem repeats at multiple loci in Mycobacterium tuberculosis. J. Clin. Microbiol. 43:5034-5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smittipat, N., and P. Palittapongarnpim. 2000. Identification of possible loci of variable number of tandem repeats in Mycobacterium tuberculosis. Tuberc. Lung Dis. 80:69-74. [DOI] [PubMed] [Google Scholar]
- 24.Supply, P., C. Allix, S. Lesjean, M. Cardoso-Oelemann, S. Rusch-Gerdes, E. Willery, E. Savine, P. de Haas, H. van Deutekom, S. Roring, P. Bifani, N. Kurepina, B. Kreiswirth, C. Sola, N. Rastogi, V. Vatin, M. C. Gutierrez, M. Fauville, S. Niemann, R. Skuce, K. Kremer, C. Locht, and D. van Soolingen. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Supply, P., J. Magdalena, S. Himpens, and C. Locht. 1997. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 26:991-1003. [DOI] [PubMed] [Google Scholar]
- 27.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 28.van Deutekom, H., S. P. Hoijng, P. E. de Haas, M. W. Langendam, A. Horsman, D. van Soolingen, and R. A. Coutinho. 2004. Clustered tuberculosis cases: do they represent recent transmission and can they be detected earlier. Am. J. Respir. Crit. Care Med. 169:806-810. [DOI] [PubMed] [Google Scholar]
- 29.van Deutekom, H., P. Supply, P. E. de Haas, E. Willery, S. P. Hoijng, C. Locht, R. A. Coutinho, and D. van Soolingen. 2005. Molecular typing of Mycobacterium tuberculosis by mycobacterial interspersed repetitive unit-variable-number tandem repeat analysis, a more accurate method for identifying epidemiological links between patients with tuberculosis. J. Clin. Microbiol. 43:4473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Soolingen, D., M. W. Borgdorff, P. E. de Haas, M. M. Sebek, J. Veen, M. Dessens, K. Kremer, and J. D. van Embden. 1999. Molecular epidemiology of tuberculosis in The Netherlands: a nationwide study from 1993 through 1997. J. Infect. Dis. 180:726-736. [DOI] [PubMed] [Google Scholar]
- 32.Warren, R. M., G. D. van der Spuy, M. Richardson, N. Beyers, C. Booysen, M. A. Behr, and P. D. van Helden. 2002. Evolution of the IS6110-based restriction fragment length polymorphism pattern during the transmission of Mycobacterium tuberculosis. J. Clin. Microbiol. 40:1277-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yaganehdoost, A., E. A. Graviss, M. W. Ross, G. J. Adams, S. Ramaswamy, A. Wanger, R. Frothingham, H. Soini, and J. M. Musser. 1999. Complex transmission dynamics of clonally related virulent Mycobacterium tuberculosis associated with barhopping by predominantly human immunodeficiency virus-positive gay men. J. Infect. Dis. 180:1245-1251. [DOI] [PubMed] [Google Scholar]
- 34.Yeh, R. W., A. Ponce de Leon, C. B. Agasino, J. A. Hahn, C. L. Daley, P. C. Hopewell, and P. M. Small. 1998. Stability of Mycobacterium tuberculosis DNA genotypes. J. Infect. Dis. 177:1107-1111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.