Abstract
A recently developed multi-virulence-locus sequence typing (MVLST) method showed improved discriminatory power for subtyping genetically diverse Listeria monocytogenes isolates and identified epidemic clone II isolates associated with two recent U.S. multistate listeriosis outbreaks. To evaluate the ability of MVLST to distinguish other epidemic clones and outbreak strains of L. monocytogenes, 58 outbreak-related isolates from 14 outbreaks and 49 unrelated isolates were analyzed. Results showed that MVLST provided very high discriminatory power (0.99), epidemiological concordance (1.0), stability, and typeability. MVLST accurately identified three previously known epidemic clones (epidemic clones I, II, and III) and redefined another epidemic clone (epidemic clone IV) in serotype 4b of L. monocytogenes. A set of 28 single nucleotide polymorphisms (SNPs) differentiated all epidemiologically unrelated isolates. A subset of 16 SNPs differentiated all epidemic clones and outbreak strains. Phylogenetic analysis showed congruence between MVLST clusters, serotypes, and previously defined genetic lineages of L. monocytogenes. SNPs in virulence genes appear to be excellent molecular markers for the epidemiological investigation of epidemics and outbreaks caused by L. monocytogenes.
Listeria monocytogenes is a gram-positive intracellular food-borne pathogen that can cause the sometimes fatal disease listeriosis among high-risk populations. It is found in a wide variety of reservoirs and sources in food-processing plants and contaminates ready-to-eat (RTE) foods such as soft cheeses, milk, deli meats, and hot dogs. Although the USDA and FDA have a zero-tolerance policy for L. monocytogenes in RTE food products, numerous listeriosis outbreaks have been reported in recent years (30). Because listeriosis has a long incubation period (3 to 60 days), it is often difficult to identify sources and routes of transmission by conventional epidemiological investigations (58). Therefore, molecular subtyping strategies targeting various genetic markers have been utilized to recognize an outbreak, match case isolates with food and environmental isolates, and discriminate between outbreak and nonoutbreak isolates (31).
Listeriosis outbreaks are usually caused by a small fraction of strains in the entire population of the species. Among 13 serotypes of L. monocytogenes, strains belonging to serotypes 4b, 1/2a, and 1/2b are associated with the vast majority of listeriosis outbreaks and sporadic cases (40). Most major outbreaks were caused by a small number of epidemic clones of L. monocytogenes, and most outbreak strains belong to serotype 4b (30). Early multilocus enzyme electrophoresis (MEE)-based subtyping (3) and restriction enzyme analysis (57) showed that strains from many different outbreaks were closely related even though those outbreaks were geographically and temporally distinct. These findings were also supported by data generated using ribotyping (12), virulence gene polymorphism analysis (59), and pulsed-field gel electrophoresis (PFGE) (4, 5, 28, 31). Kathariou (30, 31) subsequently compared those studies and defined four epidemic clones of L. monocytogenes (epidemic clone I [ECI], ECIa, ECII, and ECIII). Among these epidemic clones, ECI appears to be a cosmopolitan clonal group composed of serotype 4b strains involved in several major outbreaks including coleslaw (Nova Scotia, 1981), soft cheese (Switzerland, 1983 to 1987, and California, 1985), and pork tongue (France, 1992) outbreaks (31). Genetic markers unique to this epidemic clone were also identified (61). ECIa, another serotype 4b cluster, caused a pate outbreak (United Kingdom, 1988), a vegetable outbreak (Boston, MA, 1983), and a milk outbreak (Boston, MA, 1983). ECII is considered to be a newly emerged epidemic clone that was first observed in a U.S. multistate outbreak associated with contaminated hot dogs in 1998 and 1999 (31). Isolates in this outbreak had unique ribotype and PFGE patterns that were not identified in previous outbreaks (31). Multi-virulence-locus sequence typing (MVLST) and ECII PCR profiling were recently used to demonstrate that ECII isolates were also involved in the 2002 U.S. multistate listeriosis outbreak associated with turkey deli meat (8). Genome microarray analysis confirmed that isolates from these two outbreaks were closely related and belonged to a single clonal group (32). Genetic markers unique to this epidemic clone were also identified (14, 32). Although no routes of transmission between outbreaks within each epidemic clone were identified, the fact that various molecular subtyping methods consistently identified the epidemic clones strongly suggested that outbreaks within each epidemic clone were closely related. In contrast to ECI, ECIa, and ECII isolates, ECIII isolates are serotype 1/2a isolates associated with the hot dog (United States, 1989) and the turkey deli meat (United States, 2000) outbreaks (31). The genetic characteristics of isolates from these two outbreaks have not been extensively studied. However, they were considered to be epidemiologically related because they were found in the same food-processing plant and had identical PFGE patterns using different restriction enzymes (31). Nelson et al. (40) identified single nucleotide polymorphisms (SNPs) unique to each of the epidemic clones by comparing whole-genome sequences of representative isolates from each of the epidemic clones. Given the ubiquitous nature of L. monocytogenes in food processing plants, its ability to grow in foods at refrigeration temperatures, and the difficulty in detecting routes of transmission, it seems reasonable that previously or newly identified epidemic clones and outbreak strains will likely be implicated in future listeriosis outbreaks. Therefore, the identification and tracking of epidemic clones and outbreak strains remain critical for investigating and preventing listeriosis outbreaks.
Both DNA fragment-based and sequence-based molecular subtyping methods have been used to investigate listeriosis outbreaks. Two of the most important criteria for selecting molecular typing methods are discriminatory power (D) and epidemiological concordance (E) (46, 50). Discriminatory power describes the ability of a subtyping method to generate distinct and discrete units of information from unrelated isolates, whereas epidemiological concordance describes the ability of a typing system to correctly classify all epidemiologically related isolates from a well-described outbreak as the same clone (50). Therefore, an epidemiologically relevant subtyping method should be able to (i) cluster isolates that are epidemiologically related with a particular epidemic/outbreak and (ii) separate these isolates from those that are not related to the same epidemic/outbreak (8). Different subtyping methods target different molecular markers (i.e., housekeeping genes, virulence genes, restriction sites, and 16S and 23S RNA genes) and yield different levels of epidemiological relevance. PFGE targets variations within and between restriction sites and provides high discriminatory power; therefore, it is currently the “gold standard” method for the epidemiological investigation of foodborne pathogens of the U.S. Centers for Disease Control and Prevention (CDC) (24). Multilocus sequence typing (MLST), which targets housekeeping genes, was developed previously by Maiden et al. (37) to overcome the technical difficulties and limitations of fragment-based methods. MLST has proven to be very useful for studying the population structures of bacteria, including various foodborne pathogens (49). Recent studies have shown that antigen genes and virulence genes may provide additional genetic markers. Feavers et al. (15) supplemented MLST with antigen gene sequences to study an outbreak of meningococcal disease. A recent study of methicillin-resistant Staphylococcus aureus demonstrated that sequence variations in three virulence-associated loci could be used to identify epidemic clones and study the epidemiology of this pathogen (21). Packard et al. (44) also suggested that the sequencing of several virulence genes of Bordetella pertussis provided data that were useful for epidemiological investigations of this pathogen. An MLST scheme based solely on sequence analysis of six virulence genes, MVLST, was recently developed and used to subtype a limited number of genetically diverse isolates of L. monocytogenes (60). MVLST provided greater discriminatory power than ApaI-PFGE (60). MVLST was subsequently used to analyze isolates from two U.S. multistate listeriosis outbreaks associated with contaminated hot dogs in 1998 and 1999 and turkey deli meat in 2002 (8). In that study, MVLST demonstrated that both outbreaks were caused by one epidemic clone, ECII. However, the capacity of MVLST to differentiate multiple epidemic clones and outbreak strains of L. monocytogenes has not yet been evaluated. Therefore, the objective of the present study was to determine whether MVLST can differentiate multiple epidemic clones and outbreak strains of L. monocytogenes from a large number of listeriosis outbreaks and distinguish them from isolates that were presumably unrelated to those epidemics/outbreaks.
MATERIALS AND METHODS
Bacterial isolates and DNA extraction.
Isolate identification numbers, sources, and subtype data are given in Table 1. A total of 66 isolates were obtained from the Listeria collection at the CDC (Atlanta, GA), the International Life Sciences Institute (ILSI) North America Listeria strain collection (18) at Cornell University (Ithaca, NY), and a meat-processing plant in Oklahoma. Among these isolates, 34 outbreak isolates were involved in 11 listeriosis outbreaks, with two or more matched isolates belonging to each of the outbreaks. Isolates from these outbreaks were well characterized and identified as being involved in each outbreak by the CDC, the WHO, and Health Canada. Among them, isolates from eight outbreaks (Table 1) were designated by the ILSI as the standard set of isolates for evaluation and validation of molecular subtyping methods. Three isolates were associated with another three listeriosis outbreaks, with one isolate belonging to each of the outbreaks. The isolates from each of the outbreaks came from foods, environments, or patients. Isolates from 10 outbreaks were identified as belonging to ECI, ECIa, ECII, or ECIII in previous reports (8, 30, 31). Another 29 genetically diverse isolates without known epidemiological links to each other or to any of the outbreaks were selected and obtained from the Listeria collection at the Food Safety Laboratory at Cornell University. Most isolates were previously analyzed by serotyping and ribotyping and represented eight serotypes and three lineages of L. monocytogenes. The isolates were originally collected at different times from various sources including animals (sheep, turkey, cow, and goat), food-processing environments (equipment, drain, floor, etc.), finished food products (salmon, beef, turkey, etc.), non-food-processing environments (soil and sidewalk), and patients. Bacterial isolates were stored in tubes of 15% glycerol at −80°C and grown at 37°C overnight on Trypticase soy yeast extract agar plates (Difco Laboratories, Becton Dickinson, Sparks, MD). Cultures grown overnight were adjusted to an optical density at 650 nm of 0.2, which is equivalent to approximately 107 CFU/ml. For all isolates, bacterial genomic DNA was extracted using an UltraClean microbial DNA extraction kit (Mo Bio Laboratories, Solana Beach, CA) and stored at −20°C before use.
TABLE 1.
Sources, lineages, and subtypes of the 66 representative isolates sequenced in this study
| Group and isolate | Previous IDa | Sourcea | Lineagea | Serotypea | Ribotypea,e | MVLSTb |
|---|---|---|---|---|---|---|
| Epidemic clone I (30) | ||||||
| 1981 Canada coleslaw outbreak* (WHOc, Health Canada) | ||||||
| BL1301 | FSL J1-108 | Human | I | 4b | DUP-1038B | 20 |
| BL1302 | FSL J1-107 | Human | I | 4d | DUP-1038B | 20 |
| BL1303 | FSL J1-003 | Human | I | 4b | DUP-1038B | 20 |
| BL1304 | FSL N3-008 | Food | I | 4b | DUP-1038 | 20 |
| 1985 California soft cheese outbreak* (WHO) | ||||||
| BL1201 | FSL J1-002 | Human | I | 4b | DUP-1038B | 20 |
| BL1202 | FSL J1-119 | Human | I | 4b | DUP-1038B | 20 |
| BL1203 | FSL J1-110 | Food | I | 4b | DUP-1038B | 20 |
| 1983-1987 Switzerland soft cheese outbreak* (WHO) | ||||||
| BL1101 | FSL J1-123 | Human | I | 4b | DUP-1038B | 20 |
| BL1102 | FSL J1-126 | Human | I | 4b | DUP-1038B | 20 |
| BL1103 | FSL N3-022 | Food | I | 4b | DUP-1038B | 20 |
| Epidemic clone II (30) | ||||||
| 1998 Sara Lee hot dog outbreak (CDC) | ||||||
| BL2101 | H2444 | NAd | I | 4b | DUP-1044 | 19 |
| BL2102 | H3396 | NA | I | 4b | DUP-1044 | 19 |
| BL2103 | H6383 | NA | I | 4b | DUP-1044 | 19 |
| 2002 U.S. turkey deli outbreak (CDC) | ||||||
| BL2201 | J1838 | NA | I | 4b | DUP-1044A | 19 |
| BL2202 | J2230 | NA | I | 4b | DUP-1044A | 19 |
| BL2203 | J2685 | NA | I | 4b | DUP-1044A | 19 |
| BL2204 | J3006 | NA | I | 4b | DUP-1044A | 19 |
| BL2205 | J3033 | NA | I | 4b | DUP-1044A | 19 |
| BL2206 | J3200 | NA | I | 4b | DUP-1044A | 19 |
| BL2207 | J3238 | NA | I | 4b | DUP-1044A | 19 |
| Epidemic clone III (30) | ||||||
| 2000 U.S. turkey deli outbreak* (CDC) | ||||||
| BL3201 | FSL R2-603 | Food | II | 1/2a | DUP-1053A | 1 |
| BL3202 | FSL R2-499 | Food | II | 1/2a | DUP-1053A | 1 |
| 1989 U.S. hot dog outbreak* (WHO) | ||||||
| BL3101 | FSL N3-031 | Food | II | 1/2a | DUP-1053A | 1 |
| BL3102 | FSL J1-101 | Human | II | 1/2a | DUP-1053A | 1 |
| Epidemic clone IV (30) | ||||||
| 1989 United Kingdom pate outbreak* (WHO) | ||||||
| BL4101 | FSL J1-129 | Human | I | 4b | DUP-1042B | 21 |
| BL4102 | FSL J1-116 | Human | I | 4b | DUP-1042B | 21 |
| BL4103 | FSL N3-013 | Food | I | 4b | DUP-1042B | 21 |
| 1979 Boston vegetable outbreak | ||||||
| BL4201 | FSL J1-220 | Human | I | 4b | DUP-1042 | 21 |
| Other outbreaks | ||||||
| 1983 Boston milk outbreak | ||||||
| BL9101 | FSL J1-225 | Human | I | 4b | DUP-1042B | 31 |
| BL9102 | FSL R2-578 | Human | I | 4b | DUP-1042B | 31 |
| BL9103 | FSL R2-583 | Human | I | 4b | DUP-1042B | 31 |
| 2000 North Carolina soft cheese outbreak* (CDC) | ||||||
| BL9201 | FSL R2-500 | Food | I | 4b | DUP-1042B | 30 |
| BL9202 | FSL R2-501 | Human | I | 4b | DUP-1042B | 30 |
| 1987 Pennsylvania ice cream outbreak | ||||||
| BL9301 | FSL J1-012 | Human | I | 4b | DUP-1038B | 29 |
| 1981 United Kingdom Carlisle outbreak | ||||||
| BL9501 | FSL J1-105 | Human | II | 1/2a | DUP-1030 | 33 |
| 1994 Illinois chocolate milk outbreak* (WHO) | ||||||
| BL9401 | FSL R2-502 | Food | I | 1/2b | DUP-1051B | 32 |
| BL9402 | FSL R2-503 | Human | I | 1/2b | DUP-1051B | 32 |
| Nonoutbreak isolates | ||||||
| BL0002 | FSL N1-014 | Food | II | 1/2a | NA | 34 |
| BL0003 | FSL N1-011A | Environmental | I | 1/2b | NA | 35 |
| BL0006 | 3562 | Food environment | NA | NA | DUP-1041 | 36 |
| BL0007 | 3563 | Food environment | NA | NA | DUP-1041 | 36 |
| BL0026 | FSL J2-044 | Animal | I | 4b | DUP-1042 | 37 |
| BL0027 | FSL N3-010 | Food | I | 4b | NA | 38 |
| BL0028 | 3564 | Food environment | NA | NA | DUP-1041 | 36 |
| BL0029 | FSL F2-239 | Food | I | 1/2b | DUP-1042C | 39 |
| BL0030 | FSL F2-601 | Human sporadic | I | 4b | DUP-1042B | 40 |
| BL0031 | FSL H1-030 | Food environment | NA | NA | DUP-1045B | 41 |
| BL0032 | FSL F2-293 | Food | I | 1/2b | DUP-1031A | 42 |
| BL0033 | FSL S4-436 | Environment | NA | NA | DUP-1025A | 43 |
| BL0034 | FSL H1-051 | Food environment | NA | NA | DUP-1039C | 44 |
| BL0035 | FSL L3-151 | Food environment | NA | NA | DUP-1039A | 45 |
| BL0036 | FSL F2-032 | Food | II | 1/2a | DUP-1045B | 46 |
| BL0037 | FSL F2-373 | Food | II | 1/2a | DUP-1039C | 47 |
| BL0038 | FSL N4-588 | Environment | NA | NA | DUP-1045B | 48 |
| BL0039 | FSL R2-219 | Food environment | NA | NA | DUP-1062A | 49 |
| BL0040 | FSL F3-831 | Food environment | NA | NA | DUP-1045A | 50 |
| BL0041 | FSL S6-016 | Environment | NA | NA | NA | 51 |
| BL0042 | FSL F2-525 | Human sporadic | III | 4b | DUP-1061A | 52 |
| BL0043 | FSL F2-663 | Human sporadic | II | 1/2a | DUP-1054C | 53 |
| BL0044 | FSL C1-387 | Food | II | 1/2a | DUP-1039B | 45 |
| BL0045 | FSL F2-695 | Human sporadic | III | 4a | DUP-1061A | 54 |
| BL0046 | FSL F2-655 | Human sporadic | III | NA | DUP-1061A | 55 |
| BL0047 | 3569 | Food environment | NA | NA | DUP-1030 | 56 |
| BL0048 | 3571 | Food environment | NA | NA | DUP-1039 | 45 |
| BL0049 | 3580 | Food environment | NA | NA | DUP-1062 | 57 |
| BL0050 | 3583 | Food environment | NA | NA | DUP-1023 | 58 |
Isolates and information (sources, serotypes, ribotypes, and pulsotypes) were obtained from the Cornell Food Safety Laboratory, the U.S. CDC, and a meat-processing plant. ID, identification.
Sequence types were assigned according to methods reported previously by Zhang et al. (60).
Health agencies that characterized the outbreaks are included in parentheses following the name of the outbreaks. Isolates from the outbreaks followed by an asterisk were designated by the ILSI North America as a standard set of isolates for validating molecular subtyping methods.
NA, not available.
Ribotyping pattern designations followed by a capital letter indicate subribotype (i.e., DUP-1053A is a subribotype of ribogroup DUP-1053).
Selection of genes and intragenic targets.
Six virulence genes (prfA, inlB, inlC, dal, clpP, and lisR) were selected for MVLST analysis as described previously (60). Internal regions (ca. 500 bp) in each gene were amplified and sequenced as described below. To confirm that the original six-gene MVLST strategy accurately detected epidemic clones, intragenic regions of two additional virulence genes (inlA and actA) from 14 outbreak strains were also analyzed.
PCR amplification and purification of PCR amplicons.
PCR primers for all eight virulence genes (Table 2) were designed using Primer 3 software (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) on the basis of the published whole-genome sequence of L. monocytogenes strain F2365 and were synthesized at the Pennsylvania State University Shared Nucleic Acid Facility. All PCR amplifications were performed with a hot start (95°C for 15 min) prior to 30 cycles of amplification at 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, with a final extension step at 72°C for 7 min using QIAGEN HotStarTaq PCR kits (QIAGEN Inc., Valencia, CA) in a Mastercycler thermocycler (Eppendorf Scientific, Hamburg, Germany). To ensure accurate sequencing, PCR products were loaded onto a 2% UltraClean agarose gel (Mo Bio Laboratories) and electrophoresed at 120 V for 45 min with 0.5× Tris-borate-EDTA running buffer. DNA bands (ca. 500 bp) were then excised from the gel and purified using QIAquick gel extraction kits (QIAGEN Inc., Valencia, CA).
TABLE 2.
Primer sequences, size of fragment, and percent coverage of complete coding sequence for each gene analyzed in this studya
| Gene | Size of fragment (bp) | Coverage of complete CDS (%) | PCR and sequencing primers (5′-3′) | Reference or source |
|---|---|---|---|---|
| MVLST genes | ||||
| prfA | 469 | 65.69 | AACGGGATAAAACCAAAACCA (F) | 60 |
| TGCGATGCCACTTGAATATC (R) | ||||
| inlB | 433 | 22.87 | CATGGGAGAGTAACCCAACC (F) | 60 |
| GCGGTAACCCCTTTGTCATA (R) | ||||
| inlC | 418 | 46.91 | CGGGAATGCAATTTTTCACTA (F) | 60 |
| AACCATCTACATAACTCCCACCA (R) | ||||
| dal | 441 | 41.10 | GGTTTCTGCGTAGCCATTTT (F) | 60 |
| GGAAGGGGTCAATCCATACA (R) | ||||
| clpP | 419 | 70.18 | CCAACAGTAATTGAACAAACTAGCC (F) | 60 |
| GATCTGTATCGCGAGCAATG (R) | ||||
| lisR | 448 | 65.70 | CGGGGTAGAAGTTTGTCGTC (F) | 60 |
| ACGCATCACATACCCTGTCC (R) | ||||
| Additional virulence genes | ||||
| inlA | 458 | 19.01 | GCTTTCAGCTGGGCATAAC (F) | This study |
| ATTCATTTAGTTCCGCCTGT (R) | ||||
| actA | 582 | 32.01 | AAGAGGTAAATGCTTCGGACT (F) | This study |
| ATTCCATTTAGTTCCGCCTGT (R) |
CDS, coding sequence; F, forward; R, reverse.
DNA sequencing.
DNA cycle-sequencing reactions were performed at the Pennsylvania State University Shared Nucleic Acid Facility using an MJ Research Tetrad thermal cycler, 3′BigDye-labeled dideoxynucleotide triphosphates (v 3.1 dye terminators), and protocol 43032337 (Applied Biosystems, Foster City, CA). Cycle-sequencing reaction products were separated and analyzed on an ABI 3730XL DNA analyzer using the ABI Data Collection program (v 2.0). Data were analyzed with ABI Sequencing Analysis software (v 5.1.1). Both forward and reverse PCR primers (Table 2) were used as sequencing primers in separate runs. Ambiguous sites were sequenced at least three additional times to reduce sequencing errors.
Sequence analysis.
Sequence analysis was performed on a total of 107 isolates including 66 isolates from the present study (Table 1), 20 presumably unrelated isolates described previously by Zhang et al. (60), and 21 ECII isolates described previously by Chen et al. (8). Multiple sequence alignments were performed using molecular evolutionary genetic analysis software (MEGA version 3.0) (34). Sequence types were assigned as previously described (60). Briefly, different allelic sequences (with at least a 1-nucleotide difference) were assigned arbitrary numbers. For each isolate, the combination of six alleles defined its allelic profile, and a unique allelic profile was designated a sequence type. MEGA 3.0 was used to construct a neighbor-joining (34) tree of L. monocytogenes isolates using the number of nucleotide differences in the concatenated sequences (total of 2,628 bp) of six loci, with 1,000 bootstrap tests. An unrooted tree was constructed because a suitable outgroup that is closely related to L. monocytogenes and that contains sequences homologous to all six virulence genes is not available.
Calculation of D and E.
A total of 63 presumably unrelated isolates were used to calculate discriminatory power (D) using Simpson's index as described previously by Hunter and Gaston (26). These 63 isolates included 49 isolates without known epidemiological links and 14 isolates representing each of the 14 outbreaks. They represented different sources, serotypes, ribotypes, and lineages of L. monocytogenes. Thirty-four well-characterized and matched isolates from 11 listeriosis outbreaks were used to evaluate the epidemiological concordance of MVLST. Epidemiological concordance was calculated using the formula developed by the European Study Group on Epidemiological Markers (ESGEM) for each of the outbreaks (50).
Nucleotide sequence accession numbers.
Gene sequences were deposited into GenBank under accession numbers EF062596 to EF062801.
RESULTS
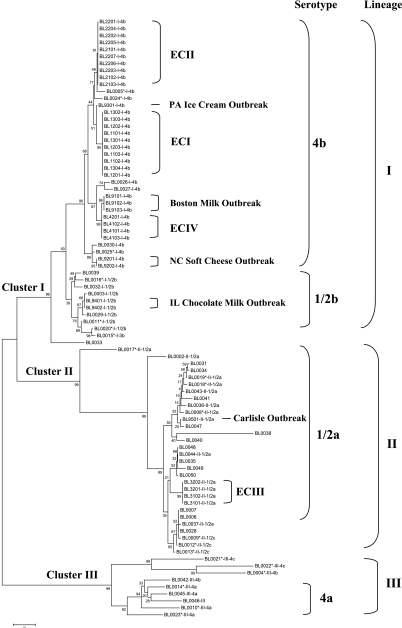
Table 1 shows the sequence types of all 66 isolates (37 outbreak isolates and 29 nonoutbreak isolates) that were sequenced in the present study. Sequence analysis was performed on MVLST data from a total of 107 isolates. Figure 1 shows the phylogenetic clustering of 86 isolates based on MVLST (66 isolates from the present study and 20 nonoutbreak isolates described previously by Zhang et al.) (60). Isolates described previously by Chen et al. (8) were found to have the sequence type that was identical to those of the newly sequenced ECII isolates and thus were not included because of limited space. A maximum parsimony tree was also constructed based on the concatenated sequences, and a similar tree topology was observed (data not shown).
FIG. 1.
Unrooted neighbor-joining tree of 86 L. monocytogenes isolates based on the number of nucleotide differences in the six MVLST virulence gene fragments analyzed. Bootstrap values (1,000 replications) are shown at the interior branches. Lineage (i.e., I, II, and III) and serotype (i.e., 4b, 1/2a, and 1/2b) information for each isolate are included after the isolate identification number. Isolates described previously by Zhang et al. (60) were assigned new BL (Borland Laboratory) identification numbers and marked with asterisks in the figure.
The 63 isolates selected for evaluating discriminatory power were representative of different serotypes, lineages, and ribotypes of L. monocytogenes and were presumed to be unrelated. MVLST identified 54 sequence types from the 63 isolates. Many epidemiologically unrelated isolates differed by only one nucleotide difference in the virulence gene sequences. Examples included BL0003 and BL9401; BL0035 and BL0050; BL0019, BL0031, and BL0034; and BL0006 and BL0037. Among the 49 isolates that were not related to the outbreaks, three isolates from the same food-processing plant (BL0006, BL0007, and BL0028) had the same MVLST sequence type, ribotype, and pulsotype. BL0035, BL0044, and BL0048 also had the same MVLST sequence type. Using Simpson's index (26), the D of MVLST was calculated to be 0.99.
Analysis of all outbreak isolates revealed that they had identical MVLST sequence types within each outbreak (Table 1). Using the formula for epidemiological concordance (E) developed by the ESGEM (50), the E for MVLST was calculated to be 1.0 for each of the outbreaks. Surprisingly, MVLST sequence types were also identical between different outbreaks within each of the three well-identified epidemic clones (ECI, ECII, and ECIII) and thus accurately clustered isolates in each epidemic clone (Table 1 and Fig. 1). The isolates from the coleslaw outbreak (Canada, 1981) and the two soft cheese outbreaks (California, 1985, and Switzerland, 1983 to 1987) had an identical sequence type (MVLST 20). The additional isolates from the hot dog outbreak (United States [multistate], 1998 to 1999) and the turkey deli meat outbreak (United States [multistate], 2002) sequenced in this study had a sequence type (MVLST 19) identical to that of the ECII isolates sequenced in the study described previously by Chen et al. (8). The isolates from the hot dog outbreak (United States, 1989) and the turkey deli outbreak (United States, 2000) also had an identical MVLST sequence type (MVLST 1). MVLST identified another epidemic cluster (MVLST 21), which included isolates from the vegetable outbreak (Boston, MA, 1979) and the pate outbreak (United Kingdom, 1989) from ECIa (Table 1). Kathariou (30) defined ECIa as including the vegetable outbreak (Boston, MA, 1979), the milk outbreak (Boston, MA, 1983), and the pate outbreak (United Kingdom, 1989) based on MEE and ribotyping (ribotype DUP-1042B) (12). However, both MEE and ribotyping have relatively low discriminatory power. For example, ribotype DUP-1042B was found in the North Carolina outbreak (BL9201 and BL9202), animal (BL 0005 and BL0025), and sporadic-case (BL0030) isolates in the present study. This ribotype was also found in various animal and environmental isolates and even in some serotype 1/2b isolates (41), and therefore, this ribotype was not unique to the three outbreaks. MVLST results showed that isolates from the Boston milk outbreak have sequence types that are different from those of the isolates from the Boston vegetable outbreak and the United Kingdom pate outbreak, and therefore, it was not considered to belong to this epidemic clone. The phylogenetic tree (Fig. 1) also revealed that ECII seemed to be more closely related to ECI than this epidemic clone. Therefore, based on our MVLST results, we renamed EC1a as ECIV and excluded the Boston milk outbreak from this epidemic clone. Isolates from the remaining five outbreaks analyzed in the present study could not be assigned to any of the four epidemic clones; therefore, they were considered single-outbreak strains or outbreak clones (Table 1 and Fig. 1).
A minimum of 28 nucleotide substitutions were identified, which provided the same discriminatory power as MVLST (D = 0.99) (Table 3). A minimum of 16 nucleotide substitutions were subsequently identified, which differentiated all epidemic clones and outbreak strains and separated them from all other unrelated isolates. These 16 nucleotide substitutions provided a discriminatory power of 0.97 and an epidemiological concordance of 1.0.
TABLE 3.
Nucleotide substitutions at the 28 nucleotide polymorphic sites in the six MVLST genes which were capable of differentiating all epidemiologically unrelated isolates analyzed in this study
| Isolate | SNP locations in the six virulence genes
|
|||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
prfAa
|
inlB
|
inlC
|
dal
|
clpP
|
lisR
|
|||||||||||||||||||||||
| * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | |||||||||||||
| 1 | 3 | 3 | 2 | 3 | 3 | 3 | 4 | 4 | 1 | 2 | 4 | 1 | 1 | 3 | 4 | 3 | 3 | 3 | 3 | 4 | ||||||||
| 8 | 2 | 6 | 5 | 9 | 2 | 1 | 7 | 7 | 2 | 2 | 1 | 4 | 7 | 6 | 4 | 1 | 9 | 5 | 5 | 0 | 4 | 8 | 1 | 2 | 6 | 8 | 3 | |
| 3 | 1 | 3 | 4 | 0 | 3 | 8 | 0 | 2 | 0 | 1 | 7 | 7 | 5 | 7 | 2 | 3 | 4 | 5 | 5 | 0 | 9 | 5 | 3 | 8 | 7 | 7 | 3 | |
| ECIb | C | C | C | G | C | G | A | G | T | T | C | C | G | G | C | T | A | G | C | A | G | C | C | G | C | T | G | C |
| ECII | . | . | . | . | . | . | . | . | . | . | . | . | T | A | . | C | . | . | . | . | . | T | . | . | . | . | . | . |
| ECIII | . | T | . | . | . | . | . | . | G | C | . | T | . | . | . | . | . | T | . | G | . | T | . | . | . | . | . | T |
| ECIV | . | . | . | . | . | . | T | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| Boston milkb | . | . | . | . | . | . | T | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . |
| NC soft cheese | T | . | . | . | T | . | . | . | . | . | . | . | T | A | . | C | . | . | . | . | . | T | . | . | . | . | . | . |
| PA ice cream | . | . | . | . | . | . | . | . | . | C | . | . | T | A | . | C | . | . | . | . | . | . | . | . | . | . | . | . |
| IL milk | . | . | . | . | T | . | . | . | G | C | . | . | T | A | . | C | . | . | . | . | . | . | . | . | . | . | . | . |
| United Kingdom Carlisle | . | . | . | A | . | A | . | . | A | C | . | . | . | . | . | . | . | T | . | G | . | T | . | . | . | . | . | . |
| BL0002 | . | T | . | . | . | . | . | . | G | C | . | . | T | A | . | C | . | T | . | G | . | T | . | A | . | . | . | T |
| BL0003 | . | . | . | . | T | . | . | . | G | . | . | . | T | A | . | C | . | . | . | . | . | . | . | . | . | G | . | T |
| BL0004 | . | T | . | . | . | . | . | . | G | . | . | . | . | . | T | . | . | . | . | . | . | . | . | A | T | . | . | . |
| BL0005 | . | . | . | . | . | . | T | A | . | . | . | . | T | A | . | C | . | . | . | . | . | T | . | . | . | . | . | . |
| BL0008 | . | T | . | A | . | . | . | . | A | C | . | . | . | . | . | . | . | T | . | G | . | T | . | . | . | . | . | . |
| BL0009 | . | T | . | A | T | . | . | . | A | C | . | T | . | . | . | . | C | T | . | G | . | T | . | . | . | . | . | T |
| BL0010 | . | T | . | . | . | . | T | A | . | . | . | . | . | . | . | C | . | . | . | . | A | . | . | A | T | . | . | T |
| BL0011 | . | . | . | . | T | . | . | . | G | C | . | . | . | A | . | C | . | . | . | . | . | T | . | . | . | . | . | T |
| BL0012 | . | T | . | A | T | . | . | . | A | C | . | T | . | . | . | . | . | T | . | . | . | T | . | . | . | . | . | . |
| BL0013 | . | T | . | A | T | . | . | . | A | C | . | . | . | . | . | . | . | T | . | G | . | T | . | . | . | . | . | . |
| BL0014 | . | T | . | . | . | . | . | A | . | . | . | . | . | . | . | C | C | . | . | . | . | . | . | A | T | . | . | . |
| BL0015 | . | . | T | . | T | . | . | . | G | C | . | . | T | A | . | C | . | . | . | . | . | T | . | . | . | . | . | . |
| BL0016 | . | . | . | . | T | . | . | . | G | C | . | . | . | . | . | C | . | . | . | . | . | T | . | . | . | . | . | . |
| BL0017 | . | T | . | . | . | A | . | . | A | C | . | . | . | . | . | . | . | . | . | G | . | T | . | . | . | . | . | . |
| BL0018 | . | T | . | A | . | . | . | . | G | C | . | T | . | . | T | . | . | T | . | G | . | T | . | . | . | . | . | . |
| BL0019 | . | T | . | A | . | A | . | . | A | C | . | T | . | . | T | . | . | T | . | G | . | T | . | . | . | . | . | . |
| BL0020 | . | . | T | . | T | . | . | . | G | C | . | . | . | A | . | C | . | . | . | . | . | T | . | . | . | . | . | . |
| BL0021 | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | T | . | . | . | . | A | T | . | . | . |
| BL0022 | . | T | . | . | . | . | . | . | G | . | . | . | . | . | . | C | . | . | . | . | . | . | . | A | T | . | . | . |
| BL0023 | . | T | . | . | . | . | . | A | . | . | . | T | . | . | T | . | . | . | . | . | A | . | . | A | T | . | . | . |
| BL0024 | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | C | . | . | . | . | . | . | . | . | . | . | . | . |
| BL0025 | . | . | . | . | T | . | . | . | . | . | . | . | T | A | . | C | . | . | . | . | A | T | . | . | . | . | . | . |
| BL0026 | . | . | . | . | . | . | T | A | . | . | T | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . |
| BL0027 | . | . | . | . | . | . | T | A | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| BL0028 | . | T | . | A | T | . | . | . | A | C | . | T | . | . | . | . | . | T | . | G | . | T | . | . | . | . | . | T |
| BL0029 | . | . | . | . | T | . | . | . | G | . | . | . | T | A | . | C | . | . | . | . | . | . | . | . | . | . | . | . |
| BL0030 | . | . | . | . | T | . | . | . | . | . | . | . | T | A | . | C | . | . | T | . | . | T | . | . | . | . | . | T |
| BL0031 | . | T | . | A | A | . | . | A | C | . | T | . | . | T | . | . | T | . | G | . | T | . | . | T | . | . | T | |
| BL0032 | . | . | . | . | T | . | . | . | G | C | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | |
| BL0033 | . | . | . | . | T | . | . | . | G | C | . | . | . | . | . | . | . | . | . | G | . | T | . | . | . | . | . | . |
| BL0034 | . | T | . | A | . | A | . | . | G | C | . | T | . | . | T | . | . | T | . | G | . | T | . | . | T | . | . | T |
| BL0035 | . | T | . | A | . | . | . | . | A | C | . | T | . | . | T | . | . | T | . | G | . | T | . | . | . | . | . | T |
| BL0036 | . | T | . | A | . | . | . | . | G | C | . | T | . | . | . | . | . | T | . | G | . | T | . | . | . | . | . | T |
| BL0037 | . | T | . | A | T | . | . | . | A | C | . | T | . | . | . | . | . | T | . | G | . | T | . | . | . | . | C | T |
| BL0038 | . | T | . | . | . | . | . | . | G | C | . | T | . | . | . | . | . | T | . | G | . | T | . | A | T | . | . | T |
| BL0039 | . | . | . | . | T | . | . | . | G | C | . | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | T |
| BL0040 | . | T | . | . | . | . | . | . | G | C | . | T | . | . | T | . | . | T | . | G | . | T | . | . | . | . | . | T |
| BL0041 | . | T | . | A | . | . | . | . | A | C | . | T | . | . | . | . | . | T | . | G | . | T | . | . | . | . | . | T |
| BL0042 | . | T | . | . | . | . | . | A | . | . | . | . | . | . | . | C | C | . | . | . | A | . | . | . | T | . | . | T |
| BL0043 | . | . | A | . | . | . | . | A | C | . | T | . | . | T | . | . | T | . | G | . | T | . | . | . | . | . | T | |
| BL0044 | . | T | . | A | . | . | . | . | A | C | . | T | . | . | T | . | . | T | . | G | . | T | . | . | . | . | . | T |
| BL0045 | . | T | . | . | . | . | . | A | . | . | . | . | . | . | . | C | C | . | . | . | A | . | . | A | T | . | . | T |
| BL0046 | . | T | . | . | . | . | T | A | . | . | . | . | . | . | . | C | C | . | . | . | A | . | . | A | T | . | . | T |
| BL0047 | . | . | A | . | A | . | . | A | C | . | T | . | . | T | . | . | T | . | G | . | T | . | . | . | . | . | T | |
| BL0048 | . | T | . | A | . | . | . | . | A | C | . | T | . | . | T | . | . | T | . | G | . | T | . | . | . | . | . | T |
| BL0049 | . | T | . | A | . | A | . | . | A | C | . | T | . | . | T | . | . | T | . | G | . | T | . | . | . | . | . | T |
| BL0050 | . | T | . | A | . | . | . | . | A | C | . | T | . | . | T | . | . | C | . | G | . | T | . | . | . | . | . | T |
Locations of the variable nucleotide sites in the virulence genes are shown by the numbers above. Position numbers that are in boldface and start with an asterisk represent the set of 16 nucleotide substitutions capable of differentiating all epidemic clones and outbreaks. Identical nucleotides at each polymorphic site are indicated by periods.
All isolates within each epidemic clone or outbreak.
To confirm that the six-gene MVLST correctly differentiated epidemic clones and outbreak strains, two additional virulence genes (inlA and actA) were selected and sequenced for isolates from all 14 outbreaks. Similar to the original six MVLST genes, all isolates within the same epidemic/outbreak had identical sequences in inlA and actA, and isolates from different epidemics/outbreaks had different inlA and actA sequences.
The neighbor-joining tree based on MVLST revealed three main clusters supported by bootstrap values of 96, 99, and 99, respectively (Fig. 1). Cluster I contained isolates from serotypes 4b, 3b, and 1/2b; cluster II contained isolates from serotypes 1/2a and 1/2c; and cluster III contained isolates from serotypes 4a, 4b, and 4c (Fig. 1). These three clusters corresponded to the three previously identified genetic lineages (I, II, and III) of L. monocytogenes (Fig. 1). MVLST results also showed congruence with serotypes of L. monocytogenes. Specifically, MVLST grouped all 56 serotype 4b isolates (except for isolates BL0004 and BL0042) into a single large cluster, as supported by a bootstrap value of 96 (Fig. 1). MVLST grouped all 1/2b isolates and a serotype 3b isolate into a single cluster, although the bootstrap value was only 39. MVLST clustered all serotype 4a isolates, although one serotype 4b isolate (BL0042) was also grouped into the same cluster (Fig. 1). MVLST did not separate serotype 1/2a and 1/2c isolates into two distinct clusters, but all serotype 1/2c isolates were in the same cluster and separated from most serotype 1/2a isolates (Fig. 1).
Sequence variations in the original six MVLST loci of all 107 L. monocytogenes isolates were determined (Table 4). The percentages of polymorphic nucleotide sites ranged from 8.10% for prfA to 19.05% for dal, and the nucleotide diversity ranged from 0.017 for prfA to 0.067 for dal. However, the six MVLST genes showed low levels of diversity among lineage I isolates. Specifically, virulence gene sequences were highly conserved among the serotype 4b isolates, with no nucleotide polymorphism observed within each epidemic clone. The percentages of polymorphic nucleotide sites ranged from 0 for clpP to 1.84% for inlB among serotype 4b outbreak isolates (Table 4). The nucleotide diversity ranged from 0 for clpP to 0.0036 for inlB. Although clpP showed no polymorphism among outbreak isolates of serotype 4b, it showed moderate polymorphism among isolates of different serotypes or lineages.
TABLE 4.
Description analysis of nucleotide sequence data in all isolates, lineage I isolates, and outbreak isolates of serotype 4b
| Isolate | Gene | No. of polymorphic sites | % of polymorphic sites | No. of synonymous mutations | No. of nonsynonymous mutations | dN/dS ratioa | π/siteb |
|---|---|---|---|---|---|---|---|
| All isolates | prfA | 38 | 8.10 | 33 | 6 | 0.015 | 0.017 |
| inlB | 53 | 12.24 | 39 | 15 | 0.080 | 0.039 | |
| inlC | 36 | 8.31 | 22 | 14 | 0.115 | 0.020 | |
| dal | 84 | 19.05 | 70 | 14 | 0.063 | 0.067 | |
| clpP | 40 | 9.55 | 38 | 5 | 0.020 | 0.028 | |
| lisR | 63 | 14.1 | 57 | 7 | 0.004 | 0.031 | |
| Lineage I isolates | prfA | 11 | 1.71 | 10 | 1 | 0.017 | 0.0033 |
| inlB | 25 | 5.77 | 20 | 5 | 0.139 | 0.023 | |
| inlC | 3 | 0.72 | 1 | 2 | 0.605 | 0.004 | |
| dal | 7 | 1.36 | 6 | 1 | 0.074 | 0.048 | |
| clpP | 15 | 3.58 | 13 | 2 | 0.067 | 0.0005 | |
| lisR | 8 | 1.79 | 5 | 3 | 0.064 | 0.004 | |
| Outbreak isolates of serotype 4b | prfA | 3 | 0.64 | 2 | 1 | 0.058 | 0.0015 |
| inlB | 8 | 1.84 | 5 | 3 | 0.377 | 0.0036 | |
| inlC | 3 | 0.72 | 1 | 2 | 0.572 | 0.0017 | |
| dal | 5 | 1.13 | 4 | 1 | 0.074 | 0.0027 | |
| clpP | 0 | 0 | 0 | 0 | 0 | ||
| lisR | 3 | 0.67 | 3 | 0 | 0.0028 |
The dN/dS ratio was not calculated when the number of nonsynonymous mutations was zero.
Average pairwise nucleotide difference per site.
DISCUSSION
A set of criteria (discriminatory power, epidemiological concordance, typeability, reproducibility, and stability) has been established to evaluate different molecular subtyping methods (1, 50, 52). D is the probability that two unrelated strains sampled from the test population will be placed into different typing groups (26). When discriminatory power is evaluated, isolates are selected to represent subtypes that are geographically, temporally, and genetically distinct from those of the entire population, which are presumably unrelated (50). A good subtyping method must provide high discriminatory power to distinguish epidemiologically unrelated isolates (19, 20). High discriminatory power is also important for studying population structures of bacterial pathogens (37). In the past two decades, researchers have targeted various genetic regions to increase the discriminatory power of subtyping methods (33, 38). However, high discriminatory power does not guarantee that the subtyping method can provide epidemiologically relevant information. For example, if a molecular subtyping method is too discriminatory, it may generate discrete subtypes among epidemiologically related isolates and thus fail to identify an ongoing outbreak. Barrett et al. (2) found that different PFGE patterns were present in a single chain of Escherichia coli transmission and suggested that the source of this outbreak would likely have been unidentified if epidemiologists had relied solely on PFGE data. Molecular subtyping methods with discriminatory power that is too high may not be useful for assessing long-term epidemiology (10), especially when the global distribution of clonal groups of infectious agents is explored, because these methods cannot establish genetic relationships and gain general insights about isolates from diverse areas (37).
Epidemiological concordance, sometimes referred to as epidemiological relevance (4, 8, 27), is another important criterion for evaluating molecular subtyping methods for epidemiological investigation. The ESGEM (50) defined epidemiological concordance as the probability that epidemiologically related isolates from presumably single-clone outbreaks are determined to be similar enough to be classified into the same clone. Therefore, the concept of epidemiological concordance was distinguished from that of discriminatory power. Epidemiological concordance drew less attention than discriminatory power in early studies of molecular subtyping (7, 16, 60) but has been emphasized recently (8, 9, 27). Hyytia-Trees et al. (27) used epidemiological relevance as an important criterion when evaluating multiple-locus variable-number tandem repeat analysis as a second-generation subtyping method for PulseNet. Those authors claimed that multiple-locus variable-number tandem repeat analysis possessed promising epidemiological relevance by correctly clustering isolates belonging to eight well-characterized Escherichia coli O157:H7 outbreaks. Chiou et al. (9) demonstrated that by successfully identifying Shigella isolates from four outbreaks, inter-IS1 spacer polymorphisms were good molecular markers for detecting and tracing the spread of Shigella sonnei strains over periods of months or years (9). Therefore, as suggested by the ESGEM (50), both high discriminatory power and high epidemiological concordance are essential criteria for molecular subtyping methods.
In the present study, MVLST sequence types were identical within each epidemic clone and outbreak strain and therefore provided data with high epidemiological concordance (E = 1.0). It is noteworthy that the sequences of all eight virulence genes (six MVLST genes plus actA and inlA) were identical within previously identified epidemic clones I, II, and III (Table 1 and Fig. 1). A review of the concepts of epidemic, outbreak, and clone suggests that MVLST results are epidemiologically meaningful. An outbreak is defined as a cluster of cases of a disease caused by a source strain in excess of what is expected during a specified period of time (46). An epidemic can be confined in time and place and represent an outbreak, be more widespread in time and place and represent an epidemic, or be a pandemic if it spreads globally (46). A clone has been defined as a group of isolates descended from a recent common ancestor which possesses similar genetic characteristics (43). Therefore, isolates that are part of the same chain of transmission within an epidemic/outbreak can be referred to as a clone (8), and according to data described previously by Wassenaar (56), these isolates often have identical or similar subtypes. Riley (46) previously proposed a working definition of a clone as a group of isolates whose discrete typing data are indistinguishable or similar by a specific subtyping method in an epidemiological setting. Based on the MVLST results in the present study, it is reasonable to define an epidemic clone of L. monocytogenes as epidemic isolates descended from a common ancestor whose virulence gene sequences remain identical over time and place. Further research is needed to determine whether or not other virulence gene sequences in L. monocytogenes are identical within epidemic clones and outbreak strains. It was actually surprising that no sequence variation was observed in the virulence loci between outbreaks spanning up to 11 years (1989 U.S. outbreak and 2000 U.S. outbreak caused by ECIII). This was especially surprising for inlA, inlB, and inlC, as they code for surface proteins, which were thought to be hypervariable (6, 40). One possible explanation for this high virulence gene sequence conservation within epidemic clones is that virulence genes carry critical functions for causing epidemics and thus may be under strong selective pressure to remain unchanged. This is supported by the low nonsynonymous to synonymous substitution (dN/dS) ratios (less than 1) of all virulence genes analyzed in this study. Merino et al. (39) identified a MutSL mismatch repair system in L. monocytogenes and demonstrated that this system kept the gene sequences and virulence highly conserved in pathogenic strains during in vivo passage in mice. Another possible reason for this lack of virulence gene sequence variation within epidemic clones is that the epidemic clones might have evolved relatively recently. Various factors may act together to cause this conservation in virulence gene sequences.
Among the four epidemic clones of L. monocytogenes, ECI and ECIV caused pandemics involving outbreaks in Europe and North America. One possible reason why epidemic clones frequently cause outbreaks is that epidemic clones may have enhanced virulence compared to isolates that are not associated with epidemics (54). If true, enhanced virulence could be caused by specific sequences in virulence genes unique to epidemic isolates (25). Although this hypothesis was tested by different researchers, no conclusive results were obtained (25, 29). Another hypothesis that may explain why certain lineages and strains cause more frequent food-borne illness is that epidemic clones may have an enhanced ability to colonize and persist in food-processing environments during interoutbreak intervals. This hypothesis is supported by the fact that ECIII isolates have been found to persist in the same food-processing plant over a period of around 11 years (30). Epidemic clones of L. monocytogenes may be transmitted to other food-processing plants by food handlers, transportation vehicles, pallets, foods, and/or food-processing equipment (54). For instance, ECII isolates may have been transmitted among meat-processing plants both between and within states in the United States (8, 22). If true, the exact mechanism(s) by which epidemic clones are transmitted between food-processing plants remains unknown but intriguing. Another possible explanation is that isolates in these epidemic clones might have a lower infectious dose than other L. monocytogenes isolates, and therefore, they are more likely to cause listeriosis when consumed.
Incorporation of nucleotide substitutions specific for epidemic clones and other outbreak isolates into an SNP genotyping strategy may allow the rapid identification of epidemic clones of L. monocytogenes. The set of 16 SNPs that were capable of differentiating all epidemic clones and outbreak strains in the present study might yield such a strategy (Table 3). Interestingly, although dal provided the largest polymorphism among all isolates, exclusion of the nucleotide substitution in dal (position 149) (Table 3) from the SNP analysis failed to differentiate only two unrelated nonoutbreak isolates (data not shown). Therefore, dal might be excluded from future MVLST-based subtyping schemes to simplify the molecular epidemiological investigation of listeriosis outbreaks.
In addition to accurately identifying all epidemic clones and outbreak strains of L. monocytogenes, MVLST clusters I, II, and III (Fig. 1) also showed congruence with the three known genetic lineages (41, 55, 59). Most serotype 4b isolates were grouped into one cluster, except for isolates BL0004 and BL0042. These two unusual serotype 4b isolates were identified as lineage III isolates, which is also in agreement with previous studies (41, 59). All serotype 1/2b isolates were also clustered into one group but with a low bootstrapping value of 39. The observed congruence between MVLST and serotypes 4b and 1/2b is consistent with the clonal population structure of L. monocytogenes lineage I isolates (41). The congruence between lineage II and lineage III serotypes and MVLST sequence types was not as great as that between MVLST results and lineage I serotypes. This is also consistent with a previous finding that lineages II and III are highly diverse, with evidence of horizontal gene transfer (41), which tends to obscure the genetic relationship suggested by sequence analysis (11). The congruence between MVLST clusters and lineages of L. monocytogenes argues that virulence gene sequences contain phylogenetic signals. The phylogenetic tree generated by MVLST provided information on the clustering of L. monocytogenes from different serotypes and lineages, as supported by high bootstrap values (Fig. 1). However, it did not provide adequate information for studying population biology, evolution history, and the genetic relationship between different lineages of L. monocytogenes because no suitable outgroup was available to root this tree.
MLST strategies utilizing housekeeping genes have been used to successfully differentiate and subtype isolates of L. monocytogenes (45, 48). MLST is also very useful for studying population genetics of bacterial species (13). MLST using housekeeping genes provided population structures that were typically more reliable than those using other genes (11) because housekeeping genes generally evolve slowly and thus are highly conserved (38). However, their utility for investigating listeriosis outbreaks remains unproven. Salcedo et al. (48) and Meinersmann et al. (38) both developed MLST schemes using housekeeping genes to subtype L. monocytogenes. They found that MLST provided satisfactory discriminatory power and reliable population structure, but they did not evaluate the epidemiological concordance of MLST using outbreak-related isolates. Revazishvili et al. (45) used four housekeeping genes and two virulence genes in an MLST scheme for subtyping of L. monocytogenes. MLST did not show good correlation with serotypes of L. monocytogenes and failed to cluster isolates within the same lineages (45). Attempts at using MLST for epidemiological typing of other pathogens have not demonstrated that it provides good epidemiological relevance. For example, Lemee et al. (35) used housekeeping genes in an MLST scheme to subtype clinical isolates of Clostridium difficile and found that MLST sometimes failed to cluster isolates from the same clinical lineages and also failed to separate isolates from different clinical lineages. Sails et al. (47) developed an MLST scheme using housekeeping genes for subtyping outbreak isolates of Campylobacter jejuni and found that MLST failed to separate some epidemiologically unrelated isolates and cluster some epidemiologically related isolates. In summary, while MLST using housekeeping genes is useful for studying population genetics, it has not been proven to provide high epidemiological relevance for investigating L. monocytogenes outbreaks. To validate this, MLST could be directly compared with MVLST using the same set of isolates analyzed in the present study.
Fragment-based subtyping methods like PFGE have greatly aided the epidemiological investigation of food-borne pathogens including L. monocytogenes. While PFGE is highly discriminatory, it does not always accurately identify the existing epidemic clones of L. monocytogenes. For example, isolates within the same epidemic clone or outbreak of L. monocytogenes did not always have an identical pulsotype. The AscI- and ApaI-PFGE patterns of some isolates from the Canada outbreak and the Switzerland outbreak differed by two to three bands, although these two outbreaks belonged to the same epidemic clone (5). In contrast, the AscI-PFGE patterns of isolates from the outbreaks in California and North Carolina differed by only one band (5), but they did not belong to the same epidemic clone (Table 1 and Fig. 1). The AscI- and ApaI-PFGE patterns of some ECII isolates from the U.S. outbreaks in 1998 to 1999 and 2002 also differed by one to two bands (8). Some other studies also showed that isolates of L. monocytogenes that have identical PFGE patterns were found to be epidemiologically unrelated (28, 36, 42). Therefore, as suggested previously by Struelens et al. (52) and Foxman et al. (17), it is sometimes difficult to correlate PFGE band differences to the genetic relatedness of different isolates. AscI-PFGE and ApaI-PFGE provided different levels of epidemiological relevance when isolates within individual outbreaks were subtyped. For example, in the Switzerland outbreak and the U.S. outbreaks in 1998 to 1999 and 2002, isolates within individual outbreaks had different ApaI-PFGE patterns (5, 8, 23), which can lead to an incorrect separation of epidemiologically related isolates from the outbreak and thus confound the epidemiology of these outbreaks (8, 32). In contrast, AscI-PFGE accurately clustered isolates within individual outbreaks and accurately separated isolates between the two outbreaks and thus was useful for short-term epidemiology in this case. Therefore, in the case of ECII outbreaks, combining MVLST (identification of the epidemic) with AscI-PFGE (separation of outbreaks within the epidemic) clarified both the long-term and short-term epidemiologies of these listeriosis outbreaks (8). Unlike PFGE, isolates within the same epidemic clone always had the same ribotypes (Table 1). However, this can be explained by the insufficient discriminatory power of ribotyping due to high conservation in 16S and 23S RNA genes (51). For example, an isolate from the ice cream outbreak in Pennsylvania had the same ribotype (DUP-1038B) as ECI outbreak isolates, but this isolate did not belong to ECI as indicated by MVLST (Fig. 1) and various other subtyping methods (30). In another study, Nightingale et al. (41) also reported that L. monocytogenes isolates with the same ribotype could be found in multiple clonal groups. Therefore, an identical ribotype does not always indicate an epidemic clone. PFGE and ribotyping both target variations within and between restriction sites that are unlikely to affect the ability of isolates to cause outbreaks. This may explain why fragment-based subtyping methods, such as PFGE and ribotyping, sometimes cannot provide high epidemiological relevance.
A basic premise of molecular subtyping is that isolates that are part of the same chain of transmission are descendants of the source strain and can be referred to as a clone (8). During the spread of the source strain or even during the isolation, passage, and storage of isolates in laboratories, isolates within the same epidemic clone may obtain minor genetic variations and thus demonstrate different subtypes (53). A good subtyping scheme should still be able to demonstrate the close relatedness of the isolates within the same epidemic clone (8). The current six-gene MVLST scheme provided subtyping data that agreed with data from other fragment-based subtyping methods and also provided very high discriminatory power (D = 0.99) and epidemiological concordance (E = 1.0). Virulence genes are probably excellent markers for investigating the molecular epidemiology of L. monocytogenes because they are critical for causing epidemics and outbreaks of listeriosis. Unlike fragment-based subtyping methods, MVLST does not need to be combined with other subtyping methods to provide accurate information on epidemic clones, serotypes, and lineages of L. monocytogenes. Furthermore, MVLST is highly stable and reproducible, and the sequence data are unambiguous and electronically portable (1). In the future, this sequence-based approach might be enhanced by including other virulence genes and/or other genes responsible for causing epidemics, such as genes that play a role in virulence gene regulation and those that help this pathogen to be transmitted to humans (8, 46). For example, genes involved in biofilm formation, competition with other organisms, resistance to cleaning and sanitizing chemicals, and growth of L. monocytogenes in RTE foods at refrigeration temperatures would be expected to enhance the ability of this pathogen to be transmitted to humans and cause disease (54). Therefore, combining virulence genes with these additional “transmission” genes might result in a multi-epidemic-locus sequence typing strategy. However, it should always be kept in mind that molecular epidemiology and conventional epidemiology must complement and support one another in order to ensure accurate epidemiology (20).
Acknowledgments
This study was supported by a U.S. Department of Agriculture Special Milk Safety grant to The Pennsylvania State University.
We thank Martin Wiedmann and Bala Swaminathan for providing some of the bacterial isolates and the ribotype, pulsotype, and other isolate information used in this research. We also thank Wyatt Lo, Natasha Brooks, and John Patton for technical assistance.
Footnotes
Published ahead of print on 10 January 2007.
REFERENCES
- 1.Aires-de-Sousa, M., K. Boye, H. de Lencastre, A. Deplano, M. C. Enright, J. Etienne, A. Friedrich, D. Harmsen, A. Holmes, X. W. Huijsdens, A. M. Kearns, A. Mellmann, H. Meugnier, J. K. Rasheed, E. Spalburg, B. Strommenger, M. J. Struelens, F. C. Tenover, J. Thomas, U. Vogel, H. Westh, J. Xu, and W. Witte. 2006. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J. Clin. Microbiol. 44:619-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett, T. J., P. Gerner-Smidt, and B. Swaminathan. 2006. Interpretation of pulsed-field gel electrophoresis patterns in foodborne disease investigations and surveillance. Foodborne Pathog. Dis. 3:20-31. [DOI] [PubMed] [Google Scholar]
- 3.Bibb, W. F., B. G. Gellin, R. Weaver, B. Schwartz, B. D. Plikaytis, M. W. Reeves, R. W. Pinner, and C. V. Broome. 1990. Analysis of clinical and food-borne isolates of Listeria monocytogenes in the United States by multilocus enzyme electrophoresis and application of the method to epidemiologic investigations. Appl. Environ. Microbiol. 56:2133-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosch, R., M. Brett, B. Catimel, J. B. Luchansky, B. Ojeniyi, and J. Rocourt. 1996. Genomic fingerprinting of 80 strains from the WHO multicenter international typing study of Listeria monocytogenes via pulsed-field gel electrophoresis (PFGE). Int. J. Food Microbiol. 32:343-355. [DOI] [PubMed] [Google Scholar]
- 5.Buchrieser, C., R. Brosch, B. Catimel, and J. Rocourt. 1993. Pulsed-field gel electrophoresis applied for comparing Listeria monocytogenes strains involved in outbreaks. Can. J. Microbiol. 39:395-401. [DOI] [PubMed] [Google Scholar]
- 6.Cabanes, D., P. Dehoux, O. Dussurget, L. Frangeul, and P. Cossart. 2002. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 10:238-245. [DOI] [PubMed] [Google Scholar]
- 7.Cai, S., D. Y. Kabuki, A. Y. Kuaye, T. G. Cargioli, M. S. Chung, R. Nielsen, and M. Wiedmann. 2002. Rational design of DNA sequence-based strategies for subtyping Listeria monocytogenes. J. Clin. Microbiol. 40:3319-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Y., W. Zhang, and S. J. Knabel. 2005. Multi-virulence-locus sequence typing clarifies epidemiology of recent listeriosis outbreaks in the United States. J. Clin. Microbiol. 43:5291-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiou, C.-S., H.-L. Wei, Y.-W. Wang, J.-C. Liao, and C.-C. Li. 2006. Usefulness of inter-IS1 spacer polymorphisms for subtyping of Shigella sonnei isolates. J. Clin. Microbiol. 44:3928-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coenye, T., and J. J. LiPuma. 2002. Multilocus restriction typing: a novel tool for studying global epidemiology of Burkholderia cepacia complex infection in cystic fibrosis. J. Infect. Dis. 185:1454-1462. [DOI] [PubMed] [Google Scholar]
- 11.Cooper, J. E., and E. J. Feil. 2004. Multilocus sequence typing—what is resolved? Trends Microbiol. 12:373-377. [DOI] [PubMed] [Google Scholar]
- 12.De Cesare, A., J. L. Bruce, T. R. Dambaugh, M. E. Guerzoni, and M. Wiedmann. 2001. Automated ribotyping using different enzymes to improve discrimination of Listeria monocytogenes isolates, with a particular focus on serotype 4b strains. J. Clin. Microbiol. 39:3002-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 14.Evans, M. R., B. Swaminathan, L. M. Graves, E. Altermann, T. R. Klaenhammer, R. C. Fink, S. Kernodle, and S. Kathariou. 2004. Genetic markers unique to Listeria monocytogenes serotype 4b differentiate epidemic clone II (hot dog outbreak strains) from other lineages. Appl. Environ. Microbiol. 70:2383-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feavers, I. M., S. J. Gray, R. Urwin, J. E. Russell, J. A. Bygraves, E. B. Kaczmarski, and M. C. Maiden. 1999. Multilocus sequence typing and antigen gene sequencing in the investigation of a meningococcal disease outbreak. J. Clin. Microbiol. 37:3883-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foley, S. L., S. Simjee, J. Meng, D. G. White, P. F. McDermott, and S. Zhao. 2004. Evaluation of molecular typing methods for Escherichia coli O157:H7 isolates from cattle, food, and humans. J. Food Prot. 67:651-657. [DOI] [PubMed] [Google Scholar]
- 17.Foxman, B., L. Zhang, J. S. Koopman, S. D. Manning, and C. F. Marrs. 2005. Choosing an appropriate bacterial typing technique for epidemiologic studies. Epidemiol. Perspect. Innov. 2:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fugett, E., E. Fortes, C. Nnoka, and M. Wiedmann. 2006. International Life Sciences Institute North America Listeria monocytogenes Strain Collection: development of standard Listeria monocytogenes strain sets for research and validation studies. J. Food Prot. 69:2929-2938. [DOI] [PubMed] [Google Scholar]
- 19.Fussing, V., K. Barfod, R. Nielsen, K. Moller, J. P. Nielsen, H. C. Wegener, and M. Bisgaard. 1998. Evaluation and application of ribotyping for epidemiological studies of Actinobacillus pleuropneumoniae in Denmark. Vet. Microbiol. 62:145-162. [DOI] [PubMed] [Google Scholar]
- 20.Gerner-Smidt, P., K. Hise, J. Kincaid, S. Hunter, S. Rolando, E. Hyytia-Trees, E. M. Ribot, and B. Swaminathan. 2006. PulseNet USA: a five-year update. Foodborne Pathog. Dis. 3:9-19. [DOI] [PubMed] [Google Scholar]
- 21.Gomes, A. R., S. Vinga, M. Zavolan, and H. de Lencastre. 2005. Analysis of the genetic variability of virulence-related loci in epidemic clones of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 49:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottlieb, S. L., E. C. Newbern, P. M. Griffin, L. M. Graves, R. M. Hoekstra, N. L. Baker, S. B. Hunter, K. G. Holt, F. Ramsey, M. Head, P. Levine, G. Johnson, D. Schoonmaker-Bopp, V. Reddy, L. Kornstein, M. Gerwel, J. Nsubuga, L. Edwards, S. Stonecipher, S. Hurd, D. Austin, M. A. Jefferson, S. D. Young, K. Hise, E. D. Chernak, and J. Sobel. 2006. Multistate outbreak of listeriosis linked to turkey deli meat and subsequent changes in US regulatory policy. Clin. Infect. Dis. 42:29-36. [DOI] [PubMed] [Google Scholar]
- 23.Graves, L. M., S. B. Hunter, A. R. Ong, D. Schoonmaker-Bopp, K. Hise, L. Kornstein, W. E. DeWitt, P. S. Hayes, E. Dunne, P. Mead, and B. Swaminathan. 2005. Microbiological aspects of the investigation that traced the 1998 outbreak of listeriosis in the United States to contaminated hot dogs and establishment of molecular subtyping-based surveillance for Listeria monocytogenes in the PulseNet network. J. Clin. Microbiol. 43:2350-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graves, L. M., and B. Swaminathan. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65:55-62. [DOI] [PubMed] [Google Scholar]
- 25.Gray, M. J., R. N. Zadoks, E. D. Fortes, B. Dogan, S. Cai, Y. Chen, V. N. Scott, D. E. Gombas, K. J. Boor, and M. Wiedmann. 2004. Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70:5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyytia-Trees, E., S. C. Smole, P. A. Fields, B. Swaminathan, and E. M. Ribot. 2006. Second generation subtyping: a proposed PulseNet protocol for multiple-locus variable-number tandem repeat analysis of Shiga toxin-producing Escherichia coli O157 (STEC O157). Foodborne Pathog. Dis. 3:118-131. [DOI] [PubMed] [Google Scholar]
- 28.Jacquet, C., B. Catimel, R. Brosch, C. Buchrieser, P. Dehaumont, V. Goulet, A. Lepoutre, P. Veit, and J. Rocourt. 1995. Investigations related to the epidemic strain involved in the French listeriosis outbreak in 1992. Appl. Environ. Microbiol. 61:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeffers, G. T., J. L. Bruce, P. L. McDonough, J. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147:1095-1104. [DOI] [PubMed] [Google Scholar]
- 30.Kathariou, S. 2003. Foodborne outbreaks of listeriosis and epidemic-associated lineages of Listeria monocytogenes. In M. E. Torrence and R. E. Isaacson (ed.), Microbial food safety in animal agriculture. Iowa State University Press, Ames, IA.
- 31.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811-1829. [DOI] [PubMed] [Google Scholar]
- 32.Kathariou, S., L. Graves, C. Buchrieser, P. Glaser, R. M. Siletzky, and B. Swaminathan. 2006. Involvement of closely related strains of a new clonal group of Listeria monocytogenes in the 1998-99 and 2002 multistate outbreaks of foodborne listeriosis in the United States. Foodborne Pathog. Dis. 3:292-302. [DOI] [PubMed] [Google Scholar]
- 33.Keys, C., S. Kemper, and P. Keim. 2005. Highly diverse variable number tandem repeat loci in the E. coli O157:H7 and O55:H7 genomes for high-resolution molecular typing. J. Appl. Microbiol. 98:928-940. [DOI] [PubMed] [Google Scholar]
- 34.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 35.Lemee, L., A. Dhalluin, M. Pestel-Caron, J. F. Lemeland, and J. L. Pons. 2004. Multilocus sequence typing analysis of human and animal Clostridium difficile isolates of various toxigenic types. J. Clin. Microbiol. 42:2609-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukinmaa, S., K. Aarnisalo, M. L. Suihko, and A. Siitonen. 2004. Diversity of Listeria monocytogenes isolates of human and food origin studied by serotyping, automated ribotyping and pulsed-field gel electrophoresis. Clin. Microbiol. Infect. 10:562-568. [DOI] [PubMed] [Google Scholar]
- 37.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meinersmann, R. J., R. W. Phillips, M. Wiedmann, and M. E. Berrang. 2004. Multilocus sequence typing of Listeria monocytogenes by use of hypervariable genes reveals clonal and recombination histories of three lineages. Appl. Environ. Microbiol. 70:2193-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merino, D., H. Reglier-Poupet, P. Berche, and A. Charbit. 2002. A hypermutator phenotype attenuates the virulence of Listeria monocytogenes in a mouse model. Mol. Microbiol. 44:877-887. [DOI] [PubMed] [Google Scholar]
- 40.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nightingale, K. K., K. Windham, and M. Wiedmann. 2005. Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J. Bacteriol. 187:5537-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okwumabua, O., M. O'Connor, E. Shull, K. Strelow, M. Hamacher, T. Kurzynski, and D. Warshauer. 2005. Characterization of Listeria monocytogenes isolates from food animal clinical cases: PFGE pattern similarity to strains from human listeriosis cases. FEMS Microbiol. Lett. 249:275-281. [DOI] [PubMed] [Google Scholar]
- 43.Orskov, F., and I. Orskov. 1983. From the National Institutes of Health. Summary of a workshop on the clone concept in the epidemiology, taxonomy, and evolution of the Enterobacteriaceae and other bacteria. J. Infect. Dis. 148:346-357. [DOI] [PubMed] [Google Scholar]
- 44.Packard, E. R., R. Parton, J. G. Coote, and N. K. Fry. 2004. Sequence variation and conservation in virulence-related genes of Bordetella pertussis isolates from the UK. J. Med. Microbiol. 53:355-365. [DOI] [PubMed] [Google Scholar]
- 45.Revazishvili, T., M. Kotetishvili, O. C. Stine, A. S. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2004. Comparative analysis of multilocus sequence typing and pulsed-field gel electrophoresis for characterizing Listeria monocytogenes strains isolated from environmental and clinical sources. J. Clin. Microbiol. 42:276-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riley, L. W. 2004. Molecular epidemiology of infectious diseases: principles and practices. ASM Press, Washington, DC.
- 47.Sails, A. D., B. Swaminathan, and P. I. Fields. 2003. Utility of multilocus sequence typing as an epidemiological tool for investigation of outbreaks of gastroenteritis caused by Campylobacter jejuni. J. Clin. Microbiol. 41:4733-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salcedo, C., L. Arreaza, B. Alcala, L. de la Fuente, and J. A. Vazquez. 2003. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 41:757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spratt, B. G., and M. C. Maiden. 1999. Bacterial population genetics, evolution and epidemiology. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354:701-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Struelens, M. J. 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2-11. [DOI] [PubMed] [Google Scholar]
- 51.Struelens, M. J. 1998. Molecular epidemiologic typing systems of bacterial pathogens: current issues and perspectives. Mem. Inst. Oswaldo Cruz 93:581-585. [DOI] [PubMed] [Google Scholar]
- 52.Struelens, M. J., Y. De Gheldre, and A. Deplano. 1998. Comparative and library epidemiological typing systems: outbreak investigations versus surveillance systems. Infect. Control Hosp. Epidemiol. 19:565-569. [DOI] [PubMed] [Google Scholar]
- 53.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tompkin, R. B. 2002. Control of Listeria monocytogenes in the food-processing environment. J. Food Prot. 65:709-725. [DOI] [PubMed] [Google Scholar]
- 55.Ward, T. J., L. Gorski, M. K. Borucki, R. E. Mandrell, J. Hutchins, and K. Pupedis. 2004. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 186:4994-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wassenaar, T. M. 2003. Molecular typing of pathogens. Berl. Munch. Tierarztl. Wochenschr. 116:447-453. [PubMed] [Google Scholar]
- 57.Wesley, I. V., and F. Ashton. 1991. Restriction enzyme analysis of Listeria monocytogenes strains associated with food-borne epidemics. Appl. Environ. Microbiol. 57:969-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiedmann, M. 2002. Molecular subtyping methods for Listeria monocytogenes. J. AOAC Int. 85:524-531. [PubMed] [Google Scholar]
- 59.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, W., B. M. Jayarao, and S. J. Knabel. 2004. Multi-virulence-locus sequence typing of Listeria monocytogenes. Appl. Environ. Microbiol. 70:913-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng, W., and S. Kathariou. 1995. Differentiation of epidemic-associated strains of Listeria monocytogenes by restriction fragment length polymorphism in a gene region essential for growth at low temperatures (4°C). Appl. Environ. Microbiol. 61:4310-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]