Abstract
Accurate determination of infection with Kaposi's sarcoma-associated herpesvirus (KSHV) has been hindered by the lack of a “gold standard” for comparison of serological assays used to estimate KSHV prevalence in serosurveys conducted in different settings. We have evaluated the performance of five in-house (developed at University College London [UCL], United Kingdom, and at the virology laboratory of the Instituto de Medicine Tropical [IMT] in Sao Paulo, Brazil) and two commercial (ABI and DIAVIR) serological assays to detect antibodies to latency-associated nuclear antigen (LANA) and to lytic KSHV antigens. We used a variety of serum samples assembled to represent populations likely to be at high, intermediate, and low risk of KSHV infection in Brazil. Composite reference standard panels were prepared based on clinical and serological parameters, against which assay performances were assessed using conventional Bayesian statistics and latent class analysis (LCA). Against the clinical reference standard, in-house immunofluorescence assays to detect anti-LANA antibodies (IFA-LANA) produced at UCL and IMT had similar performances, with sensitivities of 61% (95% confidence interval [CI], 48% to 74%) and 72% (95% CI, 58% to 83%) and specificities of 99% (95% CI, 94% to 100%) and 100% (95% CI, 96% to 100%), respectively, and only the IMT IFA-LANA was included in LCA, together with the IMT IFA-lytic and four enzyme-linked immunosorbent assays (ELISAs). The LCA indicated that the IMT whole-virus ELISA performed best (sensitivity, 87% [95% CI, 81% to 91%]; and specificity, 100% [95% CI, 98% to 100%]), confirming the results obtained with the conventional statistical approach. Commercially available ELISA-based tests yielded the lowest specificities using a spectrum of serum samples. The evaluation of KSHV serological assays is warranted before planning serosurveys in various settings.
Kaposi's sarcoma-associated herpesvirus (KSHV) was identified in 1994 as herpesvirus-like DNA particles in tumor biopsies from AIDS patients with Kaposi's sarcoma (KS) (7). Later, KSHV was recognized as the etiologic agent of all clinical forms of KS, including that occurring in elderly people from the Mediterranean and Middle East (classic KS), that affecting sub-Saharan African children (endemic KS), and that following iatrogenic immunosuppression in solid-organ transplant patients (iatrogenic KS) (10). KSHV has also been associated with two other diseases, namely, primary effusion lymphoma (5) and the human immunodeficiency virus (HIV)-associated plasmablastic cell variant of multicentric Castleman's disease (11), a B-cell lymphoproliferative disorder. KSHV-associated cancer cells express latent genes involved in disruption of cell cycle regulation. The major KSHV antigen expressed during the latent phase of infection is termed latency-associated nuclear antigen 1 (LANA-1), which is encoded by open reading frame (ORF) 73, a gene implicated in cell transformation and in inhibition of tumor suppressor p53 (14) and retinoblastoma (30) proteins. The KSHV lytic cycle can be chemically induced in latently infected body-cavity-based lymphoma 1 (BCBL-1) cell lines by treatment with tetradecanoyl phorbol ester acetate (TPA) to trigger viral replication and the production of different cytoplasmic viral lytic antigens (19).
In the absence of a culture system derived from KS tumor-associated cell lines to allow direct demonstration of KSHV (2, 18), the diagnosis of KSHV infection has relied on molecular techniques and the use of serological assays to detect antibodies to KSHV antigens produced during latent or lytic phases of the viral life cycle (13, 15, 26). The immunofluorescence assay to detect anti-LANA antibodies (IFA-LANA) has long been considered a reference serological test to detect KSHV infection (23). However, with a sensitivity as low as 64 to 67% (9, 29) among AIDS patients with KS, this first-generation assay alone is of limited use in large seroepidemiological studies. KSHV latent antigen (LANA-1) (31) and lytic products associated with ORF 65 (35) and ORF K8.1 (6) are the most immunogenic antigens described so far and have thus been included in second-generation serological assays, such as enzyme-linked immunosorbent assays (ELISAs) and Western blotting (39). However, given the great variety of antibody responses to KSHV antigens and the lack of a clearly defined “gold standard” to compare new tests, information obtained from various serosurveys can be confusing. The assessment of the performance of newly developed assays is required in order to allow the conduct and interpretation of studies regarding the epidemiology and pathogenesis of KSHV. We therefore conducted a comparative evaluation of commercial kits and in-house KSHV serological assays of the first and second generations produced in specialized virology laboratories in the United Kingdom and Brazil.
MATERIALS AND METHODS
Study population.
The study population comprised 449 serum samples obtained during cross-sectional studies conducted between 1995 and 2003 by the Virology Laboratory of the Instituto de Medicina Tropical (IMT) of the University of São Paulo, Brazil (3, 24), and stored at −20°C. Duplicate aliquots were prepared and relabeled to carry out blind and parallel testing at IMT and at the CR UK Viral Oncology Group, Wolfson Institute for Biomedical Research, University College of London (UCL), United Kingdom.
Seven groups of patients were selected based on their posterior probability of being KSHV infected or uninfected, as determined by clinical data and IFA-LANA results (Table 1). Group 1 included 57 AIDS patients with histologically proven KS; group 2 included 60 AIDS patients from São Paulo who previously tested KSHV seropositive by in-house (IMT) IFA-LANA; group 3 included 60 AIDS patients from São Paulo with unknown KSHV serostatus but an absence of KS lesions; group 4 included 60 patients attending a São Paulo clinic for sexually transmitted infections; group 5 included 47 HIV-negative men who have sex with men enrolled in a cohort study for HIV prevention in São Paulo; group 6 included 60 blood donors from Salvador, Bahia State; and group 7 included 105 infants (1 to 2 years old) from São Paulo who had participated in a measles vaccine research program.
TABLE 1.
KSHV seropositivities obtained with eight different serological assays, by population group
| Clinical group (n)a | % KSHV-positive (or equivocal) samples (n)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| IFA-LANA
|
IFA-lytic | In-house ELISAs
|
Commercial ELISAs
|
||||||
| UCL | IMT | IMT whole-virus ELISA | MAP ELISA (K8.1 and 73) | ABI ELISA* (whole virus)
|
DIAVIR† (K8.1 and 65)
|
||||
| Positive | Equivocalb | Positive | Equivocalc | ||||||
| AIDS-KS patients (57) | 61.4 (57) | 71.9 (57) | 96.5 (57) | 94.7 (57) | 94.7 (57) | 100 (57) | 0 (57) | 96.5 (57) | 1.7 (57) |
| AIDS-LANA+ patients (60) | 84.2 (57) | 100 (60) | 80.0 (60) | 88.3 (60) | 85.0 (60) | 90.0 (60) | 1.7 (60) | 85.0 (60) | 1.7 (60) |
| AIDS-non-KS patients (60) | 21.8 (55) | 23.3 (60) | 10.0 (60) | 36.2 (58) | 27.6 (58) | 63.3 (60) | 6.7 (60) | 26.7 (60) | 10 (60) |
| STI clinic patients (60) | 1.8 (54) | 3.3 (60) | 5.2 (58) | 56.9 (58) | 8.3 (60) | 41.7 (60) | 15 (60) | 73.3 (60) | 3.3 (60) |
| MSM (47) | 15.2 (46) | 10.6 (47) | 12.8 (47) | 47.4 (38) | 40.4 (47) | 36.2 (47) | 2.1 (47) | 10.6 (47) | 4.3 (47) |
| Blood donors (60) | 0.0 (59) | 1.7 (60) | 1.7 (60) | 32.1 (53) | 3.4 (59) | 18.3 (60) | 1.7 (60) | 26.6 (60) | 11.7 (60) |
| Children (105) | 1.0 (97) | 0.0 (105) | 0.0 (104) | 0.0 (105) | 20.0 (105) | 20.0 (105) | 12.4 (105) | 15.2 (105) | 1.9 (105) |
| Total (449) | 24.5 (425) | 27.4 (449) | 26.7 (446) | 45.7 (429) | 37.6 (446) | 49.7 (449) | 6.5 (449) | 45.2 (449) | 4.7 (449) |
STI, sexually transmitted infections; MSM, men who have sex with men.
ABI ELISA gave 29 equivocal results.
DIAVIR gave 21 equivocal results.
KSHV serological assays. (i) In-house IFA. (a) IMT IFA-LANA and IMT IFA-lytic.
Sera were tested at a 1:40 dilution, using noninduced and TPA-induced BCBL-1 cell lines. Punctate nuclear staining in an untreated BCBL-1 cell line was considered a positive result for LANA, whereas entire cell fluorescence in approximately 20% of TPA-treated cells was considered a positive result for lytic-phase antigens (28).
(b) UCL IFA-LANA.
UCL IFA-LANA was performed using antigens obtained from the B-cell PEL cell line (BCP-1) latently infected with KSHV. A human herpesvirus 8 (HHV-8)- and Epstein-Barr virus-negative B-cell line (Ramos) was used as a control. Sera were diluted in phosphate-buffered saline (PBS) at 1:100 and supplemented with 1% dried milk as a blocking agent. Diluted sera were applied to fixed and permeable BCP-1 cells and incubated for 45 min at 22°C. Slides were then washed in blocking buffer, and fluorescein isothiocyanate-conjugated rabbit anti-human immunoglobulin G (IgG) antibody (P0214; DAKO A/S, Denmark), diluted at 1:60 in the blocking solution, was added. Following incubation at 22°C for 22 min, slides were washed in PBS and examined under a fluorescence microscope for nuclear punctate staining (15).
(ii) In-house ELISAs. (a) IMT whole-virus ELISA.
The IMT-produced whole-virus ELISA uses a lysate of crude antigens obtained from a mixture of noninduced and TPA-induced BCBL-1 cell lines. Lytic replication was induced by treating BCBL-1 cells with 20 ng/ml of TPA (Sigma) for 5 days. The cell mix was then washed in PBS (pH 7.2) and resuspended in PBS containing sodium deoxycholate at a concentration of 1:50 (based on the initial volume). The supernatant aliquots for viral preparation were obtained after sonication and centrifugation at 3,000 rpm and stored at −80°C. Microtiter plates (Nunc Polysorp) were coated overnight with 100 μl of an optimal dilution of previously titrated antigen (1:100). The plates were washed with PBS containing 0.05% Tween 20 (PBST) and incubated for 60 min with 5% skim milk in PBS; 100 μl of serum diluted 1:200 was dispensed into each well, and the plates were incubated for 1 h at 25°C. After four washings with PBST, anti-human IgG-peroxidase conjugate (P0214; DAKO A/S, Denmark) at 1:4,000 was added to the wells and incubated at 25°C for 40 min. After an additional five washings, the plates were incubated for 10 min at room temperature with a substrate/chromogen (tetramethylbenzidine; Dade Behring). The reaction was stopped and the optical density (OD) read at 450 nm, using 620 nm as a reference wavelength.
Optimal cutoff (CO) values for this assay were evaluated using all serum samples (n = 121) from the main panel, which gave concordant results with four different serological assays (IMT IFA-LANA, IMT IFA-lytic, ABI ELISA, and DIAVIR), i.e., 64 samples were KSHV seropositive and 57 were seronegative by the four assays. The IMT whole-virus ELISA ODs were plotted in a receiver-operator characteristic curve to find the optimal CO maximizing sensitivity and specificity. For a CO of 0.204, the sensitivity and specificity were 97% and 86%, respectively.
(b) MAP ORF K8.1 and 73 ELISA.
We modified an ELISA developed and optimized elsewhere (the multiple antigen peptide ELISA [MAP ELISA]) (D. Lagos, unpublished data) for the detection of antibodies against peptide epitopes of LANA-1 (25) and K8.1 (21) synthesized into a tetrameric peptide (two copies of each epitope). Sera were diluted 1:100 in PBST solution and 5% skim milk; 100 μl of diluted serum was dispensed into each well, and the plates were incubated for 1 h at 37°C. After five washings with buffer (Dade Behring), anti-human IgG-peroxidase conjugate (P0214; DAKO A/S, Denmark) at 1:4,000 was added to the wells and incubated at 37°C for 40 min. After four further washings with buffer, 100 μl of color substrate (tetramethylbenzidine; Dade Behring) was added to the wells, and the plates were left in the dark. After 10 min, the reaction was stopped and the OD read at 452 nm, using 620 nm as a reference wavelength. The best CO values for this assay were evaluated using the same 121 serum samples which were used to evaluate the IMT whole-virus ELISA CO values. The MAP ELISA ODs were plotted in a receiver-operator characteristic curve to find the optimal cutoff value to maximize the sensitivity and specificity. At the chosen cutoff value of 0.28, the associated sensitivity was 96%, and the specificity was 98%.
(iii) Commercial ELISAs. (a) HHV-8 IgG antibody ELISA kit.
The HHV-8 IgG antibody ELISA kit (Advanced Biotechnologies Inc., Columbia, MD) is based on whole-virus lysates of supernatants obtained from KS-1 cell lines (8). Viral pellets obtained from this supernatant are subjected to centrifugation on a sucrose density gradient to obtain purified virus, and these antigens are used to coat ELISA plates. Testing was performed according to the manufacturer's instructions.
(b) DIAVIR HHV-8 peptide mix ELISA.
The DIAVIR test (Biotrim International Ltd., Dublin, Ireland) combines synthetic peptide antigens from viral proteins encoded by ORF K8.1 and ORF 65. Specific antibodies are detected by incubation with anti-human IgG-peroxidase conjugate and by substrate reaction. Testing was performed according to the manufacturer's instructions (34).
Statistical analysis. (i) CRS.
We estimated the performance of the IMT and UCL in-house IFA-LANA tests by using a restricted spectrum of sera which included samples from AIDS-KS patients (group 1), representing “likely infected” patients, as well as children (group 7), representing “likely uninfected” patients (panel A; n = 162) populations. We used panel A as a reference standard for comparison of IFA-LANA tests because AIDS-KS patients and children from areas where KSHV infection is not endemic have previously been established as reference groups to assess IFA-LANA performance (16, 23). To expand the spectrum of disease in the validation group, a composite reference standard (CRS) was formed, using the concordant results of the IMT and UCL IFA-LANA tests on the entire panel (panel B; n = 393); all remaining KSHV serological assays were compared against this composite standard. The use of CRS, the process of combining imperfect diagnostic tests and clinical definitions to form an improved comparison standard, has been recommended for the evaluation of new diagnostic assays in the absence of a “gold standard” (1). To avoid incorporation bias, which occurs when the test under evaluation is included as part of the reference standard definition, the performances of IFA-LANA tests produced at UCL and IMT were compared only against a clinical standard (panel A). In comparison with the CRS (panel B), all analyses were performed, excluding equivocal results, and then repeated, considering such results to be negative. We calculated the sensitivity, specificity, and 95% confidence intervals (95% CI) of each KSHV assay compared against either panel A or panel B by using Bayes' theorem (33).
(ii) LCA.
As part of the validation strategy, we used latent class analysis (LCA), an established method of categorical data analysis which has been used previously to validate diagnostic tests in the absence of a perfect reference (20, 32). LCA is ideally suited for this purpose because it makes no assumption about the independence of errors between the diagnostic test and the reference standard, which helps to avoid the known problem of inflated validity estimates encountered with using an imperfect reference standard (38). LCA uses a nonlinear statistical model to classify groups of subjects who have shared characteristics into latent classes (32). It achieves this by maximizing a mathematical likelihood function based on the pattern of responses of input variables (e.g., results of KSHV assays). In this study, KSHV infection was defined as the latent variable, and the results for KSHV serostatus obtained with each test (KSHV infected or uninfected) were the manifest (i.e., input) items. From the input parameters, LCA was used to calculate a posterior probability of the diagnostic class (i.e., probability of KSHV infection given a pattern of results of the test) (32). Equivocal or missing data were excluded from analysis. The goodness of fit of each model was assessed using the Pearson chi-square (χ2) test, the likelihood ratio chi-square (L2) test, Akaike information criteria, and Bayesian information criteria (32, 37). We used LatentGold (version 2.0; Statistical Innovations, Belmont, MA) (37) to perform LCA. In order to allow comparability against KSHV serological assays (i.e., a dichotomous KSHV infected/uninfected serostatus), latent classes with similar conditional probabilities (i.e., probability of being KSHV infected given that the test result is positive) for KSHV infection were collapsed into a single category. Because of identical antigen preparations and a “perfect” agreement between IFA-LANA tests from UCL and IMT (kappa = 0.88; P < 0.0001), only the IMT IFA-LANA was included in the final latent class model.
RESULTS
KSHV seropositivities by the different assays are shown in Table 1. As expected, patients with a high risk of KSHV exposure (i.e., groups 1 and 2) had the highest prevalence of detectable KSHV antibodies by all tests. Groups unlikely to be infected by KSHV (i.e., children) were identified as nonreactive by most tests, except by the commercial assays and the MAP ELISA.
Performance of KSHV serological assays against a composite reference standard.
As shown in Table 2, both IFA-LANA tests had similar performances, with low sensitivities and nearly perfect specificities, when compared against a clinically defined reference group (panel A). The performances of the five remaining assays varied when compared against the composite standard. The commercial assays (i.e., ABI ELISA and DIAVIR) were shown to have high sensitivities but poor specificities. The results did not materially change after relabeling ABI ELISA and DIAVIR equivocal results as negative (ABI ELISA sensitivity, 98% [95% CI, 93% to 100%]; specificity, 68% [95% CI, 62% to 73%]; DIAVIR sensitivity, 92% [95% CI, 84% to 96%]; specificity, 69% [95% CI, 63% to 74%]). The in-house assays (i.e., IMT IFA-lytic and MAP ELISA) were shown to be the most specific (Table 2).
TABLE 2.
Performance of KSHV serological assays in comparison with a clinically defined reference standard (panel A) and composite reference standard (panel B)
| Standard panel (n) and KSHV serological assay | No. of positive samples/no. of samples tested (%) | % Sensitivity (95% CI) | % Specificity (95% CI) |
|---|---|---|---|
| A (162)a | |||
| UCL IFA-LANA | 36/154 (23) | 61 (48-74) | 99 (94-100) |
| IMT IFA- LANA | 41/162 (25) | 72 (58-83) | 100 (96-100) |
| B (393)b | |||
| In-house assays | |||
| IFA-lytic | 106/392 (27) | 85 (77-92) | 92 (88-95) |
| IMT whole-virus ELISA | 170/375 (45) | 97 (91-99) | 72 (66-77) |
| MAP ELISA (ORFs K8.1 and 73) | 151/393 (38) | 95 (88-98) | 80 (75-84) |
| Commercial assays | |||
| ABI ELISA (whole virus) | 191/369 (52) | 99 (94-100) | 65 (59-70) |
| DIAVIR (ORFs K8.1 and 65) | 181/376 (48) | 94 (87-98) | 67 (61-72) |
AIDS-KS patients (group 1), representing “likely infected” patients, and children (group 7), representing “likely uninfected” patients, as a clinical reference. Panel A was used as a reference standard for comparison of IFA-LANA tests because AIDS-KS patients and children from areas where KSHV infection is not endemic have previously been established as reference groups for assessing IFA-LANA performance (16).
CRS, based on concordant results of the IMT and UCL IFA-LANA tests on the entire panel to derive “likely positive” and “likely negative” serological reference groups. To expand the spectrum of disease in the validation group, a CRS was formed using the concordant results of the IMT and UCL IFA-LANA tests for the entire panel.
LCA.
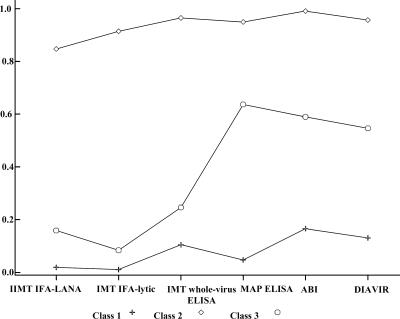
Goodness-of-fit statistics for the three LCA models considered are shown in Table 3. The best-fitting model was found to be a three-class LCA model (P = 0.5). Results for conditional probabilities derived from the best-fitting model are shown in Table 4 and Fig. 1. According to this model, samples included in class 1 had the lowest probability of having a positive test for all KSHV assays considered; therefore, we hypothesized that individuals in class 1 were the true non-KSHV-infected individuals. In contrast, individuals in class 2 had the highest probability of testing positive in all assays considered; thus, class 2 individuals were hypothesized to be the true KSHV-infected individuals. Individuals in class 3 were an intermediate case, as they had a low probability of infection based on IMT IFA-LANA, IFA-lytic, and MAP ELISA but an intermediate probability of infection by IMT whole-virus ELISA, ABI ELISA, and DIAVIR; we hypothesized that these individuals were “likely” to be KSHV infected. To allow comparability of binary KSHV assay results, classes with similar conditional probabilities of KSHV infection were used for comparison, as follows: class 2 (i.e., truly infected) and class 3 (i.e., likely infected) individuals were collapsed into a single “KSHV-positive” category, and class 1 individuals were compared as a “KSHV-negative” category.
TABLE 3.
Goodness of fit of latent class models including six KSHV serological assays (IMT IFA-LANA, IMT IFA-lytic, IMT whole-virus ELISA, MAP ELISA, ABI ELISA, and DIAVIR)
| Statistic | Value (n = 375)
|
||
|---|---|---|---|
| LC model 1 | LC model 2 | LC model 3 | |
| Degree of freedom | 57 | 50 | 43 |
| L-square value (L2)a | 985.07 | 86.22 | 38.3 |
| Chi-square value | 4,039.60 | 100.62 | 38.04 |
| P value | <0.0001 | 0.0011 | 0.5 |
| BICb | 647.24 | −210 | −216 |
| AICb | 871.07 | −13.77 | −47.68 |
| CAICb | 590.24 | −260.12 | −259.54 |
| No. (%) of samples in LC model | 166 (44) | 115 (31) | 94 (25) |
The L-square value (L2) indicates the best fit of the LC model. The larger the value, the poorer the model fits the data.
BIC, AIC, and CAIC are statistical parameters which take into account the parsimony (degrees of freedom) of the model. BIC (Bayesian information criterion), AIC (Akaike information criterion), and CAIC (consistent AIC) measure the goodness of fit of each model (the lower their values, the better the model).
TABLE 4.
Conditional probabilities derived from LC model 3, by KSHV assay
| KSHV assay and result | Conditional probability for LC model 3 (n = 375)
|
||
|---|---|---|---|
| Class 1 | Class 2 | Class 3 | |
| IMT IFA-LANA | |||
| Positive | 0.01 | 0.91 | 0.08 |
| Negative | 0.99 | 0.08 | 0.92 |
| IMT IFA-lytic | |||
| Positive | 0.02 | 0.85 | 0.16 |
| Negative | 0.98 | 0.15 | 0.84 |
| IMT whole-virus ELISA | |||
| Positive | 0.04 | 0.94 | 0.64 |
| Negative | 0.95 | 0.05 | 0.36 |
| MAP ELISA (K8.1 and 73) | |||
| Positive | 0.10 | 0.96 | 0.25 |
| Negative | 0.90 | 0.04 | 0.75 |
| ABI ELISA (whole virus) | |||
| Positive | 0.17 | 0.99 | 0.59 |
| Negative | 0.83 | 0.01 | 0.41 |
| DIAVIR (K8.1 and 65) | |||
| Positive | 0.13 | 0.96 | 0.54 |
| Negative | 0.87 | 0.04 | 0.45 |
FIG. 1.
Conditional probabilities of testing KSHV seropositive by six serological assays according to a three-class LCA model. Performances of tests are indicated as class 1 (specificity) and class 2 (sensitivity). The graph shows that samples included in class 1 had low probabilities (0.01 to 0.17) of testing positive by all assays, with IMT IFA-LANA being the test with the highest specificity. In contrast, samples included in class 2 had high probabilities (0.85 to 0.99) of testing positive by all assays, with ABI ELISA being the test with the highest sensitivity.
Comparisons of results from the best-fitting LCA model against those for each of the six serological assays are shown in Table 5. Overall, the assays had low to moderate sensitivities and high specificities. The IMT whole-virus ELISA performed best (87% sensitivity; 100% specificity), the IMT IFA-LANA was the test with the lowest sensitivity (54%) and the highest specificity (98%), and ABI ELISA was the KSHV assay with the lowest specificity (83%).
TABLE 5.
Performances of six KSHV serological assays in comparison with the latent class models presented in and Fig. 1
| KSHV assay | % Sensitivity (95% CI) | % Specificity (95% CI) |
|---|---|---|
| IMT IFA- LANA | 54 (47-61) | 98 (95-100) |
| IMT IFA-lytic | 55 (48-62) | 99 (97-100) |
| IMT whole-virus ELISA | 87 (81-91) | 100 (98-100) |
| MAP ELISA (K8.1 and 73) | 64 (57-71) | 88 (83-93) |
| ABI ELISA (whole virus) | 82 (76-87) | 83 (76-88) |
| DIAVIR (K8.1 and 65) | 79 (73-85) | 88 (82-92) |
DISCUSSION
We evaluated the performances of seven serological assays to detect KSHV antibodies to LANA and lytic antigens in a panel of Brazilian serum samples, using a combination of the classic Bayesian approach and LCA (1). A composite reference standard was defined on the basis of clinicoepidemiological data confirmed with the highly specific KSHV IFA-LANA to obtain groups with a high probability of being KSHV infected or uninfected. The use of well-characterized populations with different posterior probabilities of KSHV infection to evaluate KSHV serological assay performances has been suggested (23). In the absence of a definitive “gold standard” to determine KSHV serostatus, the combination of methodologies, by confirming the results of one approach by the other, allowed us to more reliably identify the performances of the assays under evaluation.
As expected, the in-house IFA-LANA tests yielded relatively low sensitivities and high specificities using the clinicoepidemiological reference group. A similar performance has been reported for this assay format among AIDS-KS patients (9). Other assays were able to detect KSHV infection with sensitivities ranging from 85 to 99% and specificities ranging from 65 to 92%, using the panel of sera with assorted probabilities of KSHV infection. Such variations in performance might explain the discrepant results reported in the literature when restricted spectrums of samples are used for assay evaluation. For example, the reported good performance of an ORF K8.1 ELISA (96% sensitivity and 94% specificity) using AIDS-KS patients and healthy blood donors as reference groups (21) dropped to 81% sensitivity and 86% specificity when a broader clinicoepidemiological spectrum of serum samples was included in the study. It is also possible that differences in antigen preparation might account for some of the variation in assay performance in our study. For example, MAP ELISA (ORFs K8.1 and 73) had a higher sensitivity than the IMT IFA-LANA (ORF 73 alone). This improved sensitivity with MAP ELISA could be related to the presence of latent and lytic epitopes in this assay.
Commercially available ELISAs had good sensitivities but poor specificities. These results contrast with the performance of the ABI assay in a small study of 30 AIDS-KS patients and 20 blood donors in the United States, with a reported 80% sensitivity and 100% specificity (36). Improvement in ABI ELISA specificity (>95%) at the expense of sensitivity has been reported by increasing the recommended cutoff points (4, 9, 13). Other reasons for the better performance of the ABI ELISA in other studies could be related to the use of IFA-LANA at a low, 1:20 dilution as the reference test, since low dilutions used with IFA-LANA in earlier studies were later associated with poorer specificity of this test (22).
The poorer specificity of ABI ELISA than of the IMT whole-virus ELISA, using both the CRS (65% versus 72%) and LCA (83% versus 100%), is noteworthy. Although the assays are based on the same viral target (whole virus), the observed differences might be related to antigen preparation: the ABI ELISA uses a purified virion lysate obtained from supernatant fluids from the KS-1 cell line, whereas the IMT whole-virus ELISA uses a lysate of crude antigens obtained from a mixture of noninduced and TPA-induced BCBL-1 cell lines. We did not retest equivocal results found with ABI ELISA (n = 29 [6.5%]) and DIAVIR (n = 21 [4.7%]). However, the great majority of equivocal results (28/29 for ABI ELISA and 19/21 for DIAVIR) were concentrated in groups at intermediate or low risk of KSHV infection (groups 3 to 7), and the performances of these assays did not materially change when equivocal results were eliminated or considered negative in the analysis.
We used LCA to evaluate the performances of KSHV serological assays because conventional contingency two-by-two tables might not provide accurate estimations of a test performance when no “gold standard” is available (17). LCA has already been used to assess KSHV serological assays in U.S. (27), African, and Mediterranean populations (12). In one study (12), the final LCA model indicated that the ORF K8.1 ELISA had a higher sensitivity and specificity (100% and 98%, respectively) than did ORF 65 ELISA (76% and 57%, respectively) and IFA-LANA (88% and 69%, respectively). However, no clinical or serological reference standards were used in that study to confirm the results of the LCA model. In our study, LCA provided results consistent with those obtained by conventional Bayesian methods. Firstly, the in-house IFA-LANA tests were shown to be the least sensitive, although most specific, of all assays. Secondly, the poor performances of commercial assays were confirmed. Finally, LCA showed that the IMT whole-virus ELISA had the best all-around performance. This assay thus represents a valuable alternative to commercial diagnostic tests, particularly when large numbers of samples need to be screened in resource-constrained environments. We are confident that the comparability of results between CRS and LCA strengthens the validity of our findings, since the mechanisms by which the reference standards were derived were distinct: the CRS was based on a summative approach, while LCA takes into account information from all six evaluated tests, using maximum likelihood estimation.
In summary, the lack of a “gold standard” and past performance discrepancies reported with previous KSHV serological assays prompted this thorough evaluation of available in-house and commercial tests. We recommend that testing algorithms to detect KSHV antibodies in populations should include a careful evaluation of available serological assays, using a wide spectrum of serum samples.
Acknowledgments
This work was part of the first author's Ph.D. study, which was supported by the Conselho Nacional de Desenvolvimento Científico e Technológico (CNPq), an agency of the Brazilian Ministry of Science and Technology. Financial support was also provided by the UK's Department for International Development (DFID)-funded Knowledge Programme on HIV/AIDS & STI of the London School of Hygiene & Tropical Medicine (LSHTM), United Kingdom.
We thank David Mabey and Onno Dekker (LSHTM) for their support with the study, Chris Boshoff (University College London, UCL) and Helen Weiss (LSHTM) for scientific advice, Dimitrios Lagos (UCL) for antigen preparations (MAP ELISA) and useful comments, and Dimitra Bourboulia (UCL) and Silvia Franceschi (International Agency for Research on Cancer) for useful comments and critical reviews of this paper.
Footnotes
Published ahead of print on 20 December 2006.
REFERENCES
- 1.Alonzo, T. A., and M. S. Pepe. 1999. Using a combination of reference tests to assess the accuracy of a new diagnostic test. Stat. Med. 18:2987-3003. [DOI] [PubMed] [Google Scholar]
- 2.Bubman, D., and E. Cesarman. 2003. Pathogenesis of Kaposi's sarcoma. Hematol. Oncol. Clin. N. Am. 17:717-745. [DOI] [PubMed] [Google Scholar]
- 3.Carvalho, M., S. de Carvalho, C. S. Pannuti, L. M. Sumita, and V. A. de Souza. 1998. Prevalence of herpes simplex type 2 antibodies and a clinical history of herpes in three different populations in Campinas City, Brazil. Int. J. Infect. Dis. 3:94-98. [DOI] [PubMed] [Google Scholar]
- 4.Casper, C., A. Wald, J. Pauk, S. R. Tabet, L. Corey, and C. L. Celum. 2002. Correlates of prevalent and incident Kaposi's sarcoma-associated herpesvirus infection in men who have sex with men. J. Infect. Dis. 185:990-993. [DOI] [PubMed] [Google Scholar]
- 5.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 6.Chandran, B., M. S. Smith, D. M. Koelle, L. Corey, R. Horvat, and E. Goldstein. 1998. Reactivities of human sera with human herpesvirus-8-infected BCBL-1 cells and identification of HHV-8-specific proteins and glycoproteins and the encoding cDNAs. Virology 243:208-217. [DOI] [PubMed] [Google Scholar]
- 7.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 8.Chatlynne, L. G., W. Lapps, M. Handy, Y. Q. Huang, R. Masood, A. S. Hamilton, J. W. Said, H. P. Koeffler, M. H. Kaplan, A. Friedman-Kien, P. S. Gill, J. E. Whitman, and D. V. Ablashi. 1998. Detection and titration of human herpesvirus-8-specific antibodies in sera from blood donors, acquired immunodeficiency syndrome patients, and Kaposi's sarcoma patients using a whole virus enzyme-linked immunosorbent assay. Blood 92:53-58. [PubMed] [Google Scholar]
- 9.Corchero, J. L., E. C. Mar, T. J. Spira, P. E. Pellett, and N. Inoue. 2001. Comparison of serologic assays for detection of antibodies against human herpesvirus 8. Clin. Diagn. Lab. Immunol. 8:913-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dourmishev, L. A., A. L. Dourmishev, D. Palmeri, R. A. Schwartz, and D. M. Lukac. 2003. Molecular genetics of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) epidemiology and pathogenesis. Microbiol. Mol. Biol. Rev. 67:175-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupin, N., T. L. Diss, P. Kellam, M. Tulliez, M. Q. Du, D. Sicard, R. A. Weiss, P. G. Isaacson, and C. Boshoff. 2000. HHV-8 is associated with a plasmablastic variant of Castleman disease that is linked to HHV-8-positive plasmablastic lymphoma. Blood 95:1406-1412. [PubMed] [Google Scholar]
- 12.Engels, E. A., M. D. Sinclair, R. J. Biggar, D. Whitby, P. Ebbesen, J. J. Goedert, and J. L. Gastwirth. 2000. Latent class analysis of human herpesvirus 8 assay performance and infection prevalence in sub-Saharan Africa and Malta. Int. J. Cancer 88:1003-1008. [DOI] [PubMed] [Google Scholar]
- 13.Engels, E. A., D. Whitby, P. B. Goebel, A. Stossel, D. Waters, A. Pintus, L. Contu, R. J. Biggar, and J. J. Goedert. 2000. Identifying human herpesvirus 8 infection: performance characteristics of serologic assays. J. Acquir. Immune Defic. Syndr. 23:346-354. [DOI] [PubMed] [Google Scholar]
- 14.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. P53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 15.Gambus, G., D. Bourboulia, A. Esteve, R. Lahoz, C. Rodriguez, F. Bolao, G. Sirera, R. Muga, J. del Romero, C. Boshoff, D. Whitby, and J. Casabona. 2001. Prevalence and distribution of HHV-8 in different subpopulations, with and without HIV infection, in Spain. AIDS 15:1167-1174. [DOI] [PubMed] [Google Scholar]
- 16.Gao, S. J., L. Kingsley, D. R. Hoover, T. J. Spira, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, P. Parry, Y. Chang, and P. S. Moore. 1996. Seroconversion to antibodies against Kaposi's sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi's sarcoma. N. Engl. J. Med. 335:233-241. [DOI] [PubMed] [Google Scholar]
- 17.Hawkins, D. M., J. A. Garrett, and B. Stephenson. 2001. Some issues in resolution of diagnostic tests using an imperfect gold standard. Stat. Med. 20:1987-2001. [DOI] [PubMed] [Google Scholar]
- 18.IARC. 1997. Kaposi's sarcoma herpesvirus/human herpesvirus 8. IARC Monogr. Eval. Carcinog. Risk Chem. Man 70:375-492. [PMC free article] [PubMed] [Google Scholar]
- 19.Kedes, D. H., E. Operskalski, M. Busch, R. Kohn, J. Flood, and D. Ganem. 1996. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat. Med. 2:918-924. [DOI] [PubMed] [Google Scholar]
- 20.Kim, J., D. R. Jacobs, Jr., R. V. Luepker, E. Shahar, K. L. Margolis, and M. P. Becker. 2006. Prognostic value of a novel classification scheme for heart failure: the Minnesota Heart Failure Criteria. Am. J. Epidemiol. 164:184-193. [DOI] [PubMed] [Google Scholar]
- 21.Lam, L. L., C. P. Pau, S. C. Dollard, P. E. Pellett, and T. J. Spira. 2002. Highly sensitive assay for human herpesvirus 8 antibodies that uses a multiple antigenic peptide derived from open reading frame K8.1. J. Clin. Microbiol. 40:325-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lennette, E. T., D. J. Blackbourn, and J. A. Levy. 1996. Antibodies to human herpesvirus type 8 in the general population and in Kaposi's sarcoma patients. Lancet 348:858-861. [DOI] [PubMed] [Google Scholar]
- 23.Martin, J. N., Z. Amad, C. Cossen, P. K. Lam, D. H. Kedes, K. A. Page-Shafer, D. H. Osmond, and B. Forghani. 2000. Use of epidemiologically well-defined subjects and existing immunofluorescence assays to calibrate a new enzyme immunoassay for human herpesvirus 8 antibodies. J. Clin. Microbiol. 38:696-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nascimento, M. C., L. M. Sumita, V. A. U. F. Souza, and C. S. Pannuti. 1998. Detection and direct typing of herpes simplex virus in perianal ulcers of patients with AIDS by PCR. J. Clin. Microbiol. 36:848-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen, S. J., R. Sarid, Y. Chang, and P. S. Moore. 2000. Evaluation of the latency-associated nuclear antigen (ORF73) of Kaposi's sarcoma-associated herpesvirus by peptide mapping and bacterially expressed recombinant Western blot assay. J. Infect. Dis. 182:306-310. [DOI] [PubMed] [Google Scholar]
- 26.Pellett, P. E., T. J. Spira, O. Bagasra, C. Boshoff, L. Corey, L. de Lellis, M. L. Huang, J. C. Lin, S. Matthews, P. Monini, P. Rimessi, C. Sosa, C. Wood, and J. A. Stewart. 1999. Multicenter comparison of PCR assays for detection of human herpesvirus 8 DNA in semen. J. Clin. Microbiol. 37:1298-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pellett, P. E., D. J. Wright, E. A. Engels, D. V. Ablashi, S. C. Dollard, B. Forghani, S. A. Glynn, J. J. Goedert, F. J. Jenkins, T. H. Lee, F. Neipel, D. S. Todd, D. Whitby, G. J. Nemo, and M. P. Busch. 2003. Multicenter comparison of serologic assays and estimation of human herpesvirus 8 seroprevalence among US blood donors. Transfusion 43:1260-1268. [DOI] [PubMed] [Google Scholar]
- 28.Pierrotti, L. C., L. Masami Sumita, W. Santos Freire, H. Hehl Caiaffa Filho, and V. Akico Ueda Fick de Souza. 2000. Detection of human herpesvirus 8 DNA and antibodies to latent nuclear and lytic-phase antigens in serial samples from AIDS patients with Kaposi's sarcoma. J. Clin. Virol. 16:247-251. [DOI] [PubMed] [Google Scholar]
- 29.Rabkin, C. S., T. F. Schulz, D. Whitby, E. T. Lennette, L. I. Magpantay, L. Chatlynne, and R. J. Biggar. 1998. Interassay correlation of human herpesvirus 8 serologic tests. J. Infect. Dis. 178:304-309. [DOI] [PubMed] [Google Scholar]
- 30.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 31.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S. J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rindskopf, D., and W. Rindskopf. 1986. The value of latent class analysis in medical diagnosis. Stat. Med. 5:21-27. [DOI] [PubMed] [Google Scholar]
- 33.Sacket, L., R. Haynes, G. H. Guyatt, and P. Tugwell. 1991. The interpretation of diagnostic tests, p. 69-152. In L. Sacket, R. Haynes, G. H. Guyatt, and P. Tugwell (ed.), Clinical epidemiology—a basic science for clinical medicine. Little, Brown and Company, Boston, MA.
- 34.Schatz, O., P. Monini, R. Bugarini, F. Neipel, T. F. Schulz, M. Andreoni, P. Erb, M. Eggers, J. Haas, S. Butto, M. Lukwiya, J. R. Bogner, S. Yaguboglu, J. Sheldon, L. Sarmati, F. D. Goebel, R. Hintermaier, G. Enders, N. Regamey, M. Wernli, M. Sturzl, G. Rezza, and B. Ensoli. 2001. Kaposi's sarcoma-associated herpesvirus serology in Europe and Uganda: multicentre study with multiple and novel assays. J. Med. Virol. 65:123-132. [PubMed] [Google Scholar]
- 35.Simpson, G. R., T. F. Schulz, D. Whitby, P. M. Cook, C. Boshoff, L. Rainbow, M. R. Howard, S. J. Gao, R. A. Bohenzky, P. Simmonds, C. Lee, A. De Ruiter, A. Hatzakis, R. S. Tedder, I. V. Weller, R. A. Weiss, and P. S. Moore. 1996. Prevalence of Kaposi's sarcoma associated herpesvirus infection measured by antibodies to recombinant capsid protein and latent immunofluorescence antigen. Lancet 348:1133-1138. [DOI] [PubMed] [Google Scholar]
- 36.Spira, T. J., L. Lam, S. C. Dollard, Y. X. Meng, C. P. Pau, J. B. Black, D. Burns, B. Cooper, M. Hamid, J. Huong, K. Kite-Powell, and P. E. Pellett. 2000. Comparison of serologic assays and PCR for diagnosis of human herpesvirus 8 infection. J. Clin. Microbiol. 38:2174-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vermunt, J. K., and J. Magidson. 2000. Latent gold, p. 1-80. Statistical Innovations Inc., Belmont, MA.
- 38.Walter, S. D., and L. M. Irwig. 1988. Estimation of test error rates, disease prevalence and relative risk from misclassified data: a review. J. Clin. Epidemiol. 41:923-937. [DOI] [PubMed] [Google Scholar]
- 39.Zhu, L., R. Wang, A. Sweat, E. Goldstein, R. Horvat, and B. Chandran. 1999. Comparison of human sera reactivities in immunoblots with recombinant human herpesvirus (HHV)-8 proteins associated with the latent (ORF73) and lytic (ORFs 65, K8.1A, and K8.1B) replicative cycles and in immunofluorescence assays with HHV-8-infected BCBL-1 cells. Virology 256: 381-392. [DOI] [PubMed] [Google Scholar]