Abstract
A nonfluorescent low-cost, low-density oligonucleotide array was designed for detecting the whole coronavirus genus after reverse transcription (RT)-PCR. The limit of detection was 15.7 copies/reaction. The clinical detection limit in patients with severe acute respiratory syndrome was 100 copies/sample. In 39 children suffering from coronavirus 229E, NL63, OC43, or HKU1, the sensitivity was equal to that of individual real-time RT-PCRs.
Coronaviruses (CoV) (family Coronaviridae, order Nidovirales) are large enveloped RNA viruses with a 27- to 32-kb genome of positive polarity. They comprise a very diverse spectrum of pathogens of humans and animals (2, 7). The coronavirus etiology of severe acute respiratory syndrome (SARS) and the recent discoveries of the novel human coronaviruses (hCoV) NL63 and HKU1 (5, 13, 15) have triggered intensified efforts in virus identification and diagnostics. Generic reverse transcription (RT)-PCR assays with a very broad detection range are required, but few such assays are available. None of them has been previously validated in a diagnostic setting (9, 12).
The requirement for sequencing in order to achieve strain identification limits the applicability of generic PCR assays in general. Alternative techniques, such as mass spectrometry or complex fluorescent DNA microarrays, have been proposed (10), but these will often be too sophisticated for medical facilities. We describe here a simple and feasible approach to detecting the full spectrum of coronaviruses with diagnostic sensitivity, combining generic RT-PCR and low-cost, low-density (LCD) DNA microarrays which can be read with the naked eye.
Primers for universal RT-PCR for the genus Coronavirus were designed after aligning all coronavirus RNA-dependent RNA polymerase genes. RNA-dependent RNA polymerase motifs A and C were targeted because they contain short amino acid patterns that are 100% identical in all coronaviruses (16). Primer binding regions corresponded to patterns LMGWDYPKCD and MMILSDDAV, comprising domains essential for metal ion chelation and binding of the primer 3′-end/template complex (11, 16). Reactions (25-μl mixtures) were carried out using the QIAGEN (Hilden, Germany) one-step RT-PCR kit, with 200 nM of primer PC2S2 (equimolar mixture of TTATGGGTTGGGATTATC and TGATGGGATGGGACTATC), 900 nM of primer PC2As1 (equimolar mixture of TCATCACTCAGAATCATCA, TCATCAGAAAGAATCATCA, and TCGTCGGACAAGATCATCA), 1 μl QIAGEN one-step RT-PCR kit enzyme mix, and 5 μl RNA extract. The amplification procedure comprised 30 min at 50°C; 15 min at 95°C; 10 cycles of 20 s at 94°C, 30 s starting at 62°C with a decrease of 1°C per cycle, and 40 s at 72°C; and 30 cycles of 20 s at 95°C, 30 s at 52°C, and 40 s at 72°C. To determine the sensitivity of the assay, the target regions including sufficient stretches of flanking sequence were cloned from several coronaviruses (Table 1) and transcribed into RNA (3, 4). Amplification of RNA standards yielded sensitivities in the range of single copies per assay (data not shown). However, when standards were spiked in authentic clinical samples, several log10s of sensitivity were lost. Only a nested protocol could recover sensitivity for a broad range of coronavirus RNAs. The protocol was optimized in the presence of a background of nucleic acids as encountered in routine operation. It used 1 μl of first-round PCR product, with 1× Platinum Taq buffer (Invitrogen, Karlsruhe, Germany), 200 μM deoxynucleoside triphosphate, 2.5 mM MgCl2, 80 nM of primer PCS (equimolar solutions of CTTATGGGTTGGGATTATCCTAAGTGTGA and CTTATGGGTTGGGATTATCCCAAATGTGA), 400 nM primer PCNAs (CACACAACACCTTCATCAGATAGAATCATCA), and 1 U Platinum Taq polymerase. The amplification procedure comprised 3 min at 94°C and 30 cycles of 20 s at 94°C, 30 s at 60°C, and 30 s at 72°C. To test the limit of detection of the assay and to challenge its robustness, quantified RNA transcripts were tested in the presence of high levels of background nucleic acids (human DNA, about 50 ng per reaction). As shown in Table 1, constant detection could be achieved with as little as 45 copies of RNA per reaction for all three coronavirus groups. The cumulative hit rates for all viruses were subjected to probit analysis, showing a 50% chance of detection at 15.7 copies per assay (95% confidence interval, 11 to 24 copies per assay). A 95% chance of detection required 34 copies per assay. The specificity of the assay was confirmed on samples tested in an earlier study and determined to contain influenza A virus (n = 3), influenza B virus (n = 3), human parainfluenza viruses 1 to 3 (n = 3), human metapneumovirus (n = 3), rhinoviruses (n = 3), adenoviruses (n = 3), and respiratory syncytial virus (n = 1) (8). None of these samples yielded PCR products.
TABLE 1.
Detection of quantified RNA from representative strains of all three coronavirus groups
| No. of copies per reaction | Detection of indicated virus (no. of strains correctly detected/total)a
|
|||||
|---|---|---|---|---|---|---|
| SARS-CoV | hCoV-OC43 | hCoV-229E | hCoV-NL63 | hCoV-HKU1 | AIBV | |
| 90 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| 45 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 |
| 15 | 1/3 | 1/3 | 2/3 | 0/3 | 3/3 | 0/3 |
| 5 | 0/3 | 1/3 | 1/3 | 0/3 | 3/3 | 1/3 |
| 0 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 |
Each datum point summarizes the results of three replicate tests. RNA standards were cloned and transcribed in vitro according to the following genome positions (GenBank accession numbers are shown in parentheses): SARS-CoV, positions 15008 to 15678 (NC_004718); hCoV-OC43, positions 14942 to 15606 (NC_005147); hCoV-229E, positions 14118 to 14782 (NC_002645); hCoV-NL63, positions 14037 to 14701 (AY567487), hCoV-HKU1, positions 15201 to 15865 (AY597011); avian infectious bronchitis virus (AIBV), positions 13976 to 14637 (AJ311317).
LCD arrays were established next. Oligonucleotide detection probes (see Fig. S1 in the supplemental material) were spotted on plastic microarrays using proprietary technology (Chipron, Berlin, Germany). Primer PCNAs were biotinylated and modified by use of proprietary technology (Chipron) to allow efficient hybridization. PCR products were taken directly from the tube and hybridized to LCD arrays in a 45-min procedure requiring no technical equipment except pipettes and a 37°C incubator (see Fig. S2 in the supplemental material).
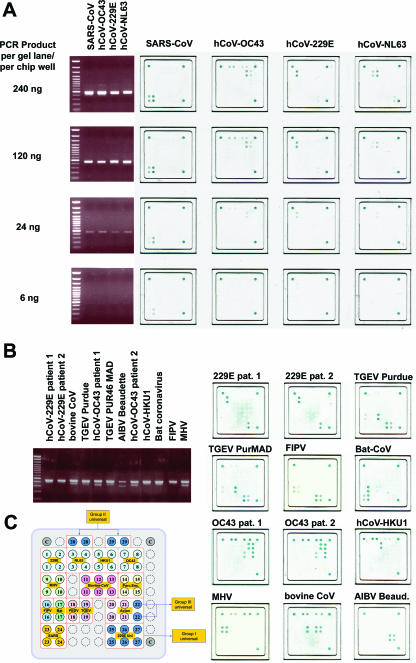
To determine whether array hybridization provided the same sensitivity as gel detection, amplification products from hCoV-229E, hCoV-NL63, hCoV-OC43, and SARS-CoV were gel purified and diluted to decreasing concentrations in PCR buffer. The same amount of PCR product was then analyzed by standard gel electrophoresis and LCD array hybridization. As shown in Fig. 1, even faint gel bands yielded corresponding hybridization signals on the array. We thus concluded that the sensitivity of array detection was comparable to that of a gel.
FIG. 1.
(A) For four different prototype coronaviruses, as indicated above the four rightmost panels, the amounts of DNA shown in the left column were subjected to gel analysis or array hybridization. Blue dots on the arrays represent hybridization signals. (B) Prototype coronaviruses as indicated above each agarose gel slot were amplified, and the depicted PCR products were hybridized to oligonucleotide arrays as shown in the panel on the right. (C) Spotting pattern of oligonucleotides on array. Each coronavirus group (I to III) is represented by a set of universal probes (blue). Strain-specific probes are depicted in different colors. Each probe is represented at least in duplicate spots. Spots in the upper left and right, as well as in the lower right corner, are staining controls containing biotin. Abbreviations: TGEV, transmissible gastroenteritis virus; AIBV, avian infectious bronchitis virus; FIPV, feline infectious peritonitis virus; MHV, mouse hepatitis virus.
It was also determined how well different coronavirus strains could be discriminated by array hybridization. RNA was extracted from cultured virus or directly from patient material, amplified, and hybridized on the array (Fig. 1). As expected, all PCRs yielded amplification products, and all PCR products gave hybridization patterns on the arrays that matched the expected virus strains. The specificity of array hybridization was confirmed by reactions carried out with mixtures containing about 0.5 μg of background DNA from human leukocytes. PCR inhibition was ruled out by spiking parallel reaction mixtures with low concentrations of feline infectious peritonitis virus or mouse hepatitis virus RNA. They did not generate any unspecific amplification signals or hybridization signals on the arrays (data not shown).
To determine a clinical limit of detection, we retested 11 original RNA preparations from throat swab samples collected from SARS patients during the 2003 epidemic. The material was tested and quantified by a commercial real-time RT-PCR assay as previously described (3). Of the 11 swabs, no detection occurred with only 3, all of which contained less than 100 copies of virus RNA per swab.
The assay was next applied to 39 stored clinical samples as summarized in Table 2. All samples had been determined in other laboratories to contain human coronaviruses by different real-time PCR protocols. Coronavirus types were determined either by sequencing or by separate, virus-specific real-time RT-PCRs (6). Because some of the samples had been stored for a long time, the material was retested in parallel with specific real-time RT-PCRs for hCoV-229E, -OC43, -HKU1, and -NL63 (6, 8). As shown in Table 2, the sensitivity of the universal coronavirus RT-PCR/LCD array was comparable to that of individual virus-specific real-time RT-PCRs. All PCR products were analyzed on LCD arrays and sequenced. All typing results were correct at the group and strain levels.
TABLE 2.
Results of real-time RT-PCR and universal coronavirus assays in stored clinical samples previously reported positive for human coronaviruses
| Assay | No. of positive results/no. of samples
|
|||
|---|---|---|---|---|
| hCoV-229E | hCoV-NL63 | hCoV-OC43 | hCoV-HKU1 | |
| Real-time RT-PCR | 3/8 | 8/12 | 11/13 | 4/6 |
| Universal coronavirus assay | 3/8 | 7/12 | 11/13 | 5/6 |
Though the genetic diversity of coronaviruses is extraordinarily high, this assay provides a simple method of detection and strain identification, obviating the need for sequencing. Its appropriateness on the clinical level has been proven by testing a large panel of virus strains as well as a sufficient number of original patient samples. Sensitivity in clinical samples ranged around 100 copies of RNA per throat swab, which is equivalent to the sensitivities of diagnostic assays which are targeted to one specific virus only, including highly optimized commercial kits (3). It is thus a suitable tool for coronavirus detection, even though enhanced anticontamination measures have to be followed to adapt the nested PCR formulation. Due to the generic features of the LCD array technology, it should be applicable to many other fields in clinical microbiology, e.g., for detecting whole virus families or for differentiating bacteria and fungi. Two examples, for genotyping Mycobacterium tuberculosis drug resistance (1) and for subtyping of human papilloma virus in squamous-cell cancer tissue (14), have recently been described. To our knowledge, this is the first application of LCD array technology to a whole viral genus. An expansion of this practical and affordable technology can be expected.
Supplementary Material
Acknowledgments
This study was supported by the German Ministry of Health as a part of funding for the National Reference Centre for Tropical Infections at the Bernhard Nocht Institute. L.K.D.S.L. is a fellow of the Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (CNPq), Brasil. C.D. is supported by the Bundesamt für Bevölkerungsschutz und Katastrophenhilfe (grant BBK-F2-440-00-167/04). L.K. is supported by grant 3200B0-113426 from the Swiss National Foundation.
Footnotes
Published ahead of print on 17 January 2007.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Aragon, L. M., F. Navarro, V. Heiser, M. Garrigo, M. Espanol, and P. Coll. 2006. Rapid detection of specific gene mutations associated with isoniazid or rifampicin resistance in Mycobacterium tuberculosis clinical isolates using non-fluorescent low-density DNA microarrays. J. Antimicrob. Chemother. 57:825-831. [DOI] [PubMed] [Google Scholar]
- 2.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 3.Drosten, C., L. L. Chiu, M. Panning, H. N. Leong, W. Preiser, J. S. Tam, S. Gunther, S. Kramme, P. Emmerich, W. L. Ng, H. Schmitz, and E. S. Koay. 2004. Evaluation of advanced reverse transcription-PCR assays and an alternative PCR target region for detection of severe acute respiratory syndrome-associated coronavirus. J. Clin. Microbiol. 42:2043-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drosten, C., S. Gottig, S. Schilling, M. Asper, M. Panning, H. Schmitz, and S. Gunther. 2002. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J. Clin. Microbiol. 40:2323-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fouchier, R. A., N. G. Hartwig, T. M. Bestebroer, B. Niemeyer, J. C. de Jong, J. H. Simon, and A. D. Osterhaus. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. USA 101:6212-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garbino, J., S. Crespo, J. D. Aubert, T. Rochat, B. Ninet, C. Deffernez, W. Wunderli, J. C. Pache, P. M. Soccal, and L. Kaiser. 2006. A prospective hospital-based study of the clinical impact of non-severe acute respiratory syndrome (non-SARS)-related human coronavirus infection. Clin. Infect. Dis. 43:1009-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez, J. M., P. Gomez-Puertas, D. Cavanagh, A. E. Gorbalenya, and L. Enjuanes. 2003. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 148:2207-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luna, L., M. Panning, K. Grywna, S. Pfefferle, and C. Drosten. 2007. Spectrum of viruses and atypical bacteria in intercontinental air travellers with symptoms of acute respiratory infection. J. Infect. Dis. 195:675-679. [DOI] [PMC free article] [PubMed]
- 9.Moes, E., L. Vijgen, E. Keyaerts, K. Zlateva, S. Li, P. Maes, K. Pyrc, B. Berkhout, L. van der Hoek, and M. Van Ranst. 2005. A novel pancoronavirus RT-PCR assay: frequent detection of human coronavirus NL63 in children hospitalized with respiratory tract infections in Belgium. BMC Infect. Dis. 5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sampath, R., S. A. Hofstadler, L. B. Blyn, M. W. Eshoo, T. A. Hall, C. Massire, H. M. Levene, J. C. Hannis, P. M. Harrell, B. Neuman, M. J. Buchmeier, Y. Jiang, R. Ranken, J. J. Drader, V. Samant, R. H. Griffey, J. A. McNeil, S. T. Crooke, and D. J. Ecker. 2005. Rapid identification of emerging pathogens: coronavirus. Emerg. Infect. Dis. 11:373-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephensen, C. B., D. B. Casebolt, and N. N. Gangopadhyay. 1999. Phylogenetic analysis of a highly conserved region of the polymerase gene from 11 coronaviruses and development of a consensus polymerase chain reaction assay. Virus Res. 60:181-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Will, C., C. Schewe, K. Schluns, and I. Petersen. 2006. HPV typing and CGH analysis for the differentiation of primary and metastatic squamous cell carcinomas of the aerodigestive tract. Cell Oncol. 28:97-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woo, P. C., S. K. Lau, C. M. Chu, K. H. Chan, H. W. Tsoi, Y. Huang, B. H. Wong, R. W. Poon, J. J. Cai, W. K. Luk, L. L. Poon, S. S. Wong, Y. Guan, J. S. Peiris, and K. Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu, X., Y. Liu, S. Weiss, E. Arnold, S. G. Sarafianos, and J. Ding. 2003. Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design. Nucleic Acids Res. 31:7117-7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.