Abstract
Using both a mass spectrometry-based method and the classical method of cloning and sequencing, we demonstrated weekly changes in the hypervariable region 1 quasispecies of a chimpanzee infected with an infectious clone, coinciding with neutralizing antibody emergence. We also used the mass spectrometry method in the clinical follow-up of a chronically infected patient over a 5-year period.
The hepatitis C virus (HCV) exists within a host as a heterogeneous population of closely related mutants, a “quasispecies.” The most studied domain of the HCV genome, in term of quasispecies, has been hypervariable region 1 (HVR1), consisting of 27 amino acids amino terminal to the E2 glycoprotein. Mutations in HVR1 often cause changes in the predicted secondary structure (17). A high rate of mutation in HVR1 likely occurs in part because it is not essential (10) and, since it contains neutralizing epitopes (8), because of immune selective pressure (13, 17, 19). Some HVR1 variants have been shown to be immune escape mutants (4, 5, 16). Studies have shown that a decrease in the diversity of quasispecies in HVR1 at the amino acid level at the time of seroconversion was predictive of viral eradication (7) and a decrease in diversity during interferon treatment was predictive of a sustained response (9). Among patients with progressive hepatitis, the rate of nonsynonymous mutations in HVR1 was higher than among patients with resolving hepatitis; synonymous mutation rates were comparable (7). These observations highlight the significance of nonsynonymous mutations.
We previously described a method to assess the changes in HVR1 amino acid sequences of mutants by in vitro translation of the amplicon mix and analysis of the resulting peptide mix by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry. This method is less costly and faster than the classical method of cloning and sequencing multiple clones (7, 9). Other rapid methods, such as single-strand conformation polymorphism and heteroduplex assays, may not distinguish between synonymous and nonsynonymous mutations. In this study, we used the MALDI-TOF method to follow HVR1 quasispecies evolution in an experimentally infected chimpanzee and applied it to the clinical follow-up of an immunosuppressed patient with chronic hepatitis C virus.
Chimpanzee 1530 was infected by intrahepatic inoculation of RNA transcripts from an infectious clone of HCV genotype 1a strain H77 (20). The housing, maintenance, and care of the chimpanzee met and exceeded all relevant guidelines and requirements. Reverse transcription-PCR of HVR1 for cloning and sequencing analysis was carried out as described previously (3, 7). Reverse transcription-PCR and MALDI-TOF analysis of HVR1 were done as described previously (1). Three different in vitro translations were done for each PCR; the molecular mass for each peptide was taken as the average ± standard deviation. Secondary structures of HVR1 were calculated from the sequences of HVR1 and the E2 glycoprotein (H77 strain) by using the Garnier-Osguthorpe-Robson (GOR) method (12), version IV (11), as implemented for GOR secondary structure prediction (http://abs.cit.nih.gov/gor/).
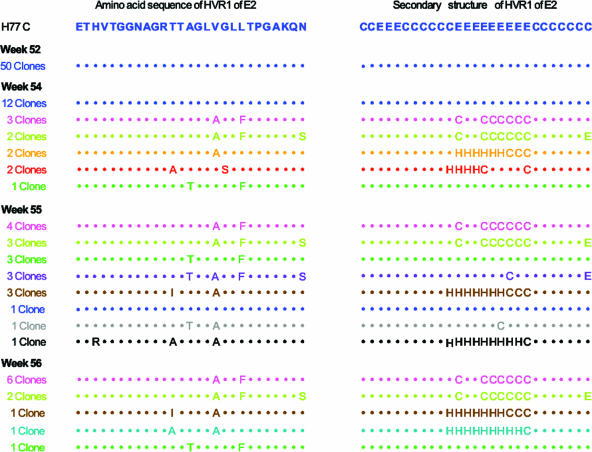
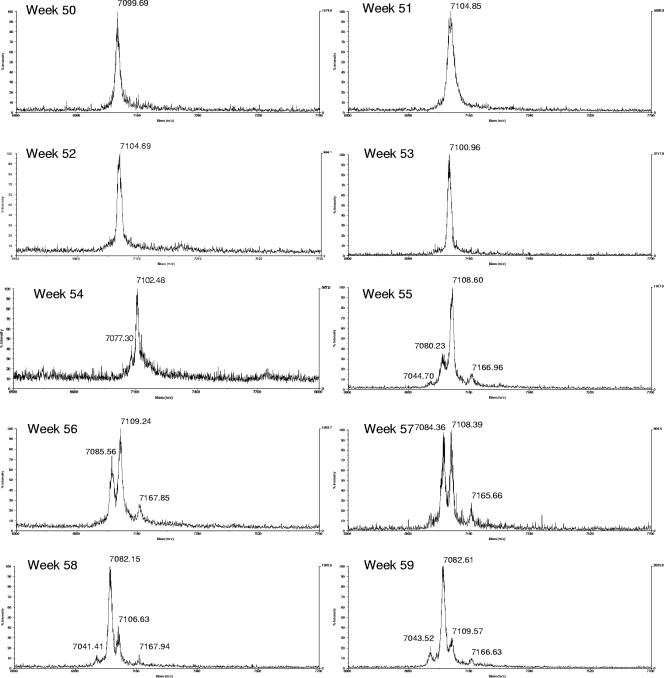
The HCV population remained clonal for about 1 year. Beginning at week 54, new sequences appeared and the HVR1 quasispecies changed very rapidly (Fig. 1). Weekly samples collected during weeks 50 to 59 were analyzed by MALDI-TOF (Fig. 2). By this method, it was found that the earliest detectable changes occurred at week 54, and by week 55 and onward, the original clone was no longer detectable (Fig. 1 and 2). In Table 1, the predicted molecular masses of the peptides, calculated as described previously (1), are compared to the observed masses. Previous estimates (1) showed that a mutant must have a frequency of more than 5% to be identified by MALDI-TOF. Because of stochastic variations, mutants present at a low frequency may not be detected by both methods (Table 1), although overall cloning and sequencing identified more mutants. It is noteworthy that by both methods, HVR1 quasispecies, originally a monoclonal population, underwent a rapid diversification, with disappearance of the original clone. This occurred at a time when the viral load was in fact decreasing (see Fig. 2A in reference 14). It is remarkable that the rapid variation in HVR1 quasispecies occurred exactly at the time when an antibody response against the original HVR1 peptide sequence was peaking (as measured by an enzyme immunoassay using a synthetic peptide corresponding to the original HVR1 sequence [14]). This anti-HVR1 response also coincided with the appearance of neutralizing antibodies against the original virus (14). These observations support the concepts that some HVR1 mutants are immune escape mutants and that quasispecies changes in HVR1 are one of the mechanisms by which HCV maintains a chronic infection. The changes in the amino acid sequences were also associated with significant changes in the predicted secondary structure in almost all mutants (Fig. 1). Changes in sequence and predicted secondary structure have previously been reported in association with immune escape mutants (17). The idea that the secondary structure is correlated with epitope recognition has been invoked for other viruses e.g., parainfluenza virus 3 (18).
FIG. 1.
Characterization of HVR1 quasispecies by cloning and sequencing of the amplicons in chimpanzee 1530. The amino acid sequences, compared to that of the original clone, are displayed in the first column. The predicted secondary structures (calculated with the GOR IV method) are displayed in the second column. H, helical (α-helix); E, extended (β-sheets); C, coil.
FIG. 2.
Weekly changes in HVR1 quasispecies in chimpanzee 1530 observed by mass spectrometry measurements of translated amplicons.
TABLE 1.
Comparison of molecular masses calculated from sequences and observed by mass spectrometrya
| Wk | Predicted molecular mass (Da) (cloning frequency) | Observed molecular mass (Da) |
|---|---|---|
| 52 | 7,101.5 (50/50) A | 7,102.85 ± 1.60 A |
| 54 | 7,101.5 (12/22) A | 7,103.33 ± 0.90 A |
| 7,107.5 (3/22) B | 7,075.26 ± 3.33 C | |
| 7,080.5 (2/22) | ||
| 7,073.5 (2/22) C | ||
| 7,101.5 (2/22) | ||
| 7,165.6 (1/22) | ||
| 55 | 7,107.5 (4/19) B | 7,106.98 ± 2.77 B |
| 7,080.5 (3/19) | 7,082.17 ± 5.68 E | |
| 7,165.6 (3/19) D | 7,166.30 ± 2.38 D | |
| 7,110.5 (3/19) | 7,048.26 ± 5.03 F | |
| 7,085.5 (3/19) E | ||
| 7,101.5 (1/19) | ||
| 7,103.5 (1/19) | ||
| 7,062.5 (1/19) | ||
| 56 | 7,107.5 (6/11) B | 7,107.14 ± 2.08 B |
| 7,080.5 (2/11) | 7,083.99 ± 3.39 E | |
| 7,085.5 (1/11) E | 7,167.75 ± 0.98 D | |
| 7,043.4 (1/11) F | ||
| 7,165.6 (1/11) D | ||
| 59 | ND | 7,111.46 ± 2.58 |
| 7,085.44 ± 2.99 E | ||
| 7,047.31 ± 3.98 F | ||
| 7,168.73 ± 2.03 D |
For the calculation of molecular masses, the amino acid sequence in Fig. 1 was used as well as the amino acid sequence encoded by the primers; posttranslational modifications were taken into account. For details, see reference 1. For the observed masses, three measurements were taken. Results are given as means ± standard deviations. Tentative associations between clones identified by the two methods are indicated by the same letter, taking into account the precision of the MALDI-TOF method (1) and the clone frequency. ND, not done.
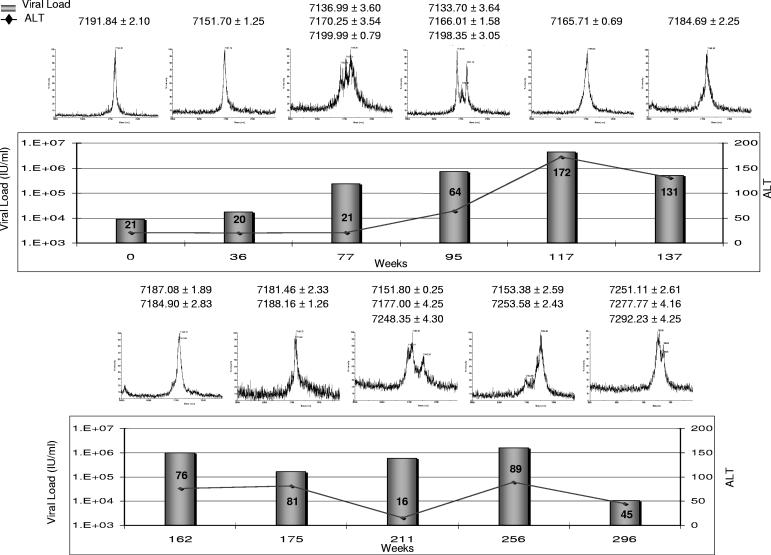
The MALDI-TOF method was also applied to the clinical follow-up of a pediatric immunosuppressed patient chronically infected with HCV genotype 1a (genotype determined as described previously [2, 15]). The study was approved by the Research Ethics Board of The Hospital for Sick Children, and the patient and his legal guardians consented to the publication of the results. Longitudinal observations integrating HCV viral load (measured using the bDNA assay Bayer VERSANT HCV RNA 3.0), alanine aminotransferase (ALT) levels, and quasispecies in HVR1 are displayed in Fig. 3; the patient underwent heart transplantation at week 0. The clinical evolution initially showed a rise in viral load along with diversification of an originally very homogeneous HCV population. A first inflammatory flare-up, indicated by a rise in ALT levels, around week 117 coincided with “pruning” of the quasispecies and was followed by a decrease in viral load and the normalization of ALT levels. A subsequent increase in viral load was also accompanied by quasispecies diversification; a second flare-up in ALT levels was accompanied by a decrease in viral load and a change in quasispecies, resulting in mutants not previously observed. These findings are consistent with the hypothesis that HVR1 diversification is followed by immune reaction (presumably mostly humoral) targeted at new mutants, resulting in inflammation and pruning of the quasispecies. This is consistent with observations by Farci et al. (6), who showed that among vertically infected children, elevated ALT levels were associated with a mono- or oligoclonal viral population, whereas in children with normal or slightly elevated ALT levels, the viral quasispecies was shown to gradually diversify. In our patient, where the immune response is blunted, the increase in viral replication may play a greater role in viral evolution. Nonetheless, episodes of inflammation still coincided with the elimination of some HVR1 mutants and a decrease in viral load. Currently, attempts at therapy for this patient would not be justified, because treatment regimens that include alpha interferon cannot be used in this clinical situation. In general, however, clinical monitoring of HVR1 quasispecies could help in choosing an optimal time for therapy initiation. As noted by Farci et al., a reduction in diversity was consistently associated with sustained viral clearance (9). It is therefore tempting to speculate that initiating therapy at a time of low diversity may further improve treatment efficacy. Because of its practicality, the MALDI-TOF-based method could therefore play a helpful role in the monitoring and treatment of patients.
FIG. 3.
Longitudinal monitoring of a patient with chronic hepatitis C virus, including ALT levels, viral load, and quasispecies monitoring.
Acknowledgments
This research was supported by a grant from the Canadian Institutes of Health Research and by the Intramural Research Program of the National Institutes of Health.
Footnotes
Published ahead of print on 17 January 2007.
REFERENCES
- 1.Ayers, M., K. Siu, E. Roberts, A. M. Garvin, and R. Tellier. 2002. Characterization of hepatitis C virus quasispecies by matrix-assisted laser desorption ionization-time of flight (mass spectrometry) mutation detection. J. Clin. Microbiol. 40:3455-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corbet, S., J. Bukh, A. Heinsen, and A. Fomsgaard. 2003. Hepatitis C virus subtyping by a core-envelope 1-based reverse transcriptase PCR assay with sequencing and its use in determining subtype distribution among Danish patients. J. Clin. Microbiol. 41:1091-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farci, P., H. J. Alter, S. Govindarajan, D. C. Wong, R. Engle, R. R. Lesniewski, I. K. Mushahwar, S. M. Desai, R. H. Miller, N. Ogata, et al. 1992. Lack of protective immunity against reinfection with hepatitis C virus. Science 258:135-140. [DOI] [PubMed] [Google Scholar]
- 4.Farci, P., H. J. Alter, D. C. Wong, R. H. Miller, S. Govindarajan, R. Engle, M. Shapiro, and R. H. Purcell. 1994. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc. Natl. Acad. Sci. USA 91:7792-7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farci, P., and R. H. Purcell. 2000. Clinical significance of hepatitis C virus genotypes and quasispecies. Semin. Liver Dis. 20:103-126. [PubMed] [Google Scholar]
- 6.Farci, P., I. Quinti, S. Farci, H. J. Alter, R. Strazzera, E. Palomba, A. Coiana, D. Cao, A. M. Casadei, R. Ledda, R. Iorio, A. Vegnente, G. Diaz, and P. A. Tovo. 2006. Evolution of hepatitis C viral quasispecies and hepatic injury in perinatally infected children followed prospectively. Proc. Natl. Acad. Sci. USA 103:8475-8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farci, P., A. Shimoda, A. Coiana, G. Diaz, G. Peddis, J. C. Melpolder, A. Strazzera, D. Y. Chien, S. J. Munoz, A. Balestrieri, R. H. Purcell, and H. J. Alter. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 8.Farci, P., A. Shimoda, D. Wong, T. Cabezon, D. De Gioannis, A. Strazzera, Y. Shimizu, M. Shapiro, H. J. Alter, and R. H. Purcell. 1996. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. USA 93:15394-15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farci, P., R. Strazzera, H. J. Alter, S. Farci, D. Degioannis, A. Coiana, G. Peddis, F. Usai, G. Serra, L. Chessa, G. Diaz, A. Balestrieri, and R. H. Purcell. 2002. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc. Natl. Acad. Sci. USA 99:3081-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forns, X., R. Thimme, S. Govindarajan, S. U. Emerson, R. H. Purcell, F. V. Chisari, and J. Bukh. 2000. Hepatitis C virus lacking the hypervariable region 1 of the second envelope protein is infectious and causes acute resolving or persistent infection in chimpanzees. Proc. Natl. Acad. Sci. USA 97:13318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garnier, J., J. F. Gibrat, and B. Robson. 1996. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 266:540-553. [DOI] [PubMed] [Google Scholar]
- 12.Garnier, J., D. J. Osguthorpe, and B. Robson. 1978. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J. Mol. Biol. 120:97-120. [DOI] [PubMed] [Google Scholar]
- 13.Kato, N., H. Sekiya, Y. Ootsuyama, T. Nakazawa, M. Hijikata, S. Ohkoshi, and K. Shimotohno. 1993. Humoral immune response to hypervariable region 1 of the putative envelope glycoprotein (gp70) of hepatitis C virus. J. Virol. 67:3923-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meunier, J. C., R. E. Engle, K. Faulk, M. Zhao, B. Bartosch, H. Alter, S. U. Emerson, F. L. Cosset, R. H. Purcell, and J. Bukh. 2005. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc. Natl. Acad. Sci. USA 102:4560-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ray, S. C., R. R. Arthur, A. Carella, J. Bukh, and D. L. Thomas. 2000. Genetic epidemiology of hepatitis C virus throughout Egypt. J. Infect. Dis. 182:698-707. [DOI] [PubMed] [Google Scholar]
- 16.Shimizu, Y. K., M. Hijikata, A. Iwamoto, H. J. Alter, R. H. Purcell, and H. Yoshikura. 1994. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J. Virol. 68:1494-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taniguchi, S., H. Okamoto, M. Sakamoto, M. Kojima, F. Tsuda, T. Tanaka, E. Munekata, E. E. Muchmore, D. A. Peterson, and S. Mishiro. 1993. A structurally flexible and antigenically variable N-terminal domain of the hepatitis C virus E2/NS1 protein: implication for an escape from antibody. Virology 195:297-301. [DOI] [PubMed] [Google Scholar]
- 18.van Wyke Coelingh, K. L., C. C. Winter, E. D. Jorgensen, and B. R. Murphy. 1987. Antigenic and structural properties of the hemagglutinin-neuraminidase glycoprotein of human parainfluenza virus type 3: sequence analysis of variants selected with monoclonal antibodies which inhibit infectivity, hemagglutination, and neuraminidase activities. J. Virol. 61:1473-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiner, A. J., H. M. Geysen, C. Christopherson, J. E. Hall, T. J. Mason, G. Saracco, F. Bonino, K. Crawford, C. D. Marion, K. A. Crawford, et al. 1992. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc. Natl. Acad. Sci. USA 89:3468-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 94:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]