Abstract
“Staphylococcus pettenkoferi” (proposed name) was identified as an unusual agent of osteomyelitis in a diabetic foot infection. The phenotypical tests used failed to give a good identification. Molecular 16S rRNA gene and rpoB sequencing allowed us to correctly identify this new species of coagulase-negative staphylococcus responsible for this chronic infection.
CASE REPORT
A 63-year-old man, diabetic since age 27 and monitored for chronic diabetic foot infection for 2 years, was admitted in the orthopedic ward of the University Regional Hospital Center, Lille, France, for disarticulation of the Chopart joint. Because of the worsening of the local septic signs (pain, redness, wound exudate, and skin necrosis) in this neuropathic patient, a transtarsal amputation was performed. X rays of the joint region confirmed the diagnosis of osteomyelitis. Laboratory data included a blood leukocyte count of 9.9 × 109/liter, with 80% of the leukocytes being polymorphonuclear; an erythrocyte sedimentation rate of 105 mm/h; and a C-reactive protein level of 18 mg/liter. During the amputation, six bone biopsy samples were cultured. As the patient was apyretic, no blood cultures were performed. The patient was successfully treated with pristinamycin for 14 weeks, giving satisfactory biological, radiological, and functional results.
Among the six bone biopsy sample cultures, four grew only a gram-positive coccus after 5 days of incubation. The presence of this bacterium in four of the major preoperational samples led us to think that the bacterium was the origin of the bone infection. The unpigmented colonies were able to grow as well in an aerobic atmosphere as in an anaerobic atmosphere. The strain grew better on Columbia agar than on other media. The isolate was catalase positive, cytochrome oxidase negative, and novobiocin susceptible. Results from testing for the presence of the clumping factor by using the Staph aureus assay (Fumouze Diagnostics, Levallois-Perret, France) and the presence of coagulase by using rabbit plasma for the coagulase test (Bio-Rad, Marnes la Coquette, France) were negative. These characteristics led us towards the Staphylococcus genus. Whatever the VITEK 2 identification cards used (fluorimetric or colorimetric cards; bioMérieux, Marcy-L'Etoile, France), the isolate could not be identified. The API ID-32 Staph strip (bioMérieux) gave a final profile number of 040010000, with low discrimination between Kocuria rosea (probability, 94.8%; typicity, 0.90) and Micrococcus lylae (probability, 5.2%; typicity, 0.70). Tests for only two reactions (acid production from d-fructose and nitrate reduction) were positive, whereas other biochemical tests (those for urease activity and pyrrolidonyl arylamidase production of acid from other sugars) were negative. Another API ID-32 Staph strip test was performed with a denser inoculum (1.0 McFarland standard) and a longer incubation time (48 h), allowing us to obtain positive results for acid production from glucose, d-fructose, and saccharose and for nitrate reduction. The API strip identified the strain with a doubtful profile (0600110200) as Staphylococcus capitis (probability, 71.5%; typicity, 0.76) or Staphylococcus auricularis (probability, 28.2%; typicity, 0.64). In vitro susceptibility tests using the VITEK 2 system showed that this bacterium was resistant only to penicillin G, ofloxacin, and fosfomycin.
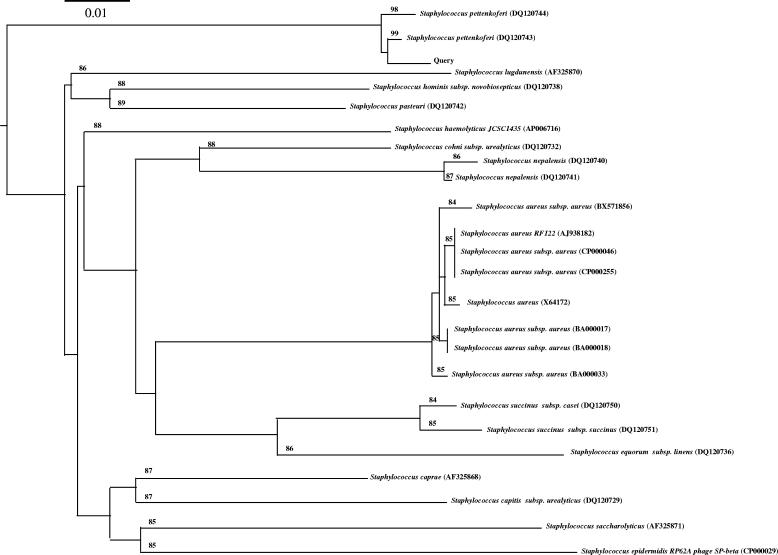
To better identify this isolate, DNA sequencing of 16S rRNA genes was performed using the MicroSeq 500 16S rRNA gene bacterial sequencing kit (Applied Biosystems, Foster City, CA). The 520-bp fragment obtained was compared with NCBI GenBank entries by using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/BLAST) and showed 99% identity with sequences from “Staphylococcus pettenkoferi” (proposed name) strain B3117 (GenBank accession number AF322022) and S. pettenkoferi strain A6664 (GenBank accession number DQ538520) previously determined by Trülzsch et al. (16). As the next-best choice, the fragment showed 96% identity with a sequence from Staphylococcus cohnii (GenBank accession number AJ717378). These results were confirmed by using bioinformatic bacterial identification (BIBI) analysis (http://pbil.univ-lyon1.fr/bibi/query.php) (data not shown). The lack of specificity observed with the 16S rRNA genes prompted us to use another target like rpoB with primers described by Drancourt and Raoult (4). The results obtained with 477 bp showed 99% identity with sequences from S. pettenkoferi strains 6664 (GenBank accession number DQ120743) and B3117 (GenBank accession number DQ120744) in the NCBI GenBank. The next-best choices were sequences from a cluster of other coagulase-negative staphylococci with a level of identity inferior to 89%. These results were confirmed by using the BIBI bank, with a phylogenetic tree presented in Fig. 1. These findings led us to identify the clinical isolate without any doubt as S. pettenkoferi.
FIG. 1.
A phylogenetic tree of the isolated rpoB sequence (query) was obtained with the BIBI program. Genetic distance is indicated on the scale (the scale bar represents 1 nucleotide substitution per 100 nucleotides). GenBank accession numbers of the rpoB sequences are shown in parentheses. Bootstrap values are indicated on the phylogenetic tree.
As reported by Cunha et al. (3), some staphylococcus strains have often been identified as K. rosea or M. lylae with the VITEK 2 system. In our case, whatever the identification cards used, either fluorimetric or colorimetric cards, the VITEK 2 system gave no identification whereas the API ID-32 Staph strip identified the isolate with weak discrimination as either K. rosea or M. lylae. For uncommon staphylococcus species, such as S. pettenkoferi, it is not surprising that the VITEK 2 system gives no identification due to the absence of these species from the database. In the case of doubtful or no identification, the identity of an isolate should be determined by either PCR (7, 9) or DNA sequencing of 16S rRNA genes (MicroSeq 500; Applied Biosystems, Foster City, CA) (1, 2, 5, 13). Sequencing of other targets such as sodA (12, 15) or rpoB (10, 11) genes may be required. In the case of a new species, the isolate would be stored for further identification. In the present work, due to the 16S rRNA gene sequences available in GenBank, the isolate was identified successfully as S. pettenkoferi. Moreover, the rpoB target was more discriminatory for the identification of S. pettenkoferi, as for the identification of other staphylococcus species as described previously by Drancourt and Raoult (4).
The present paper describes the third case of infection due to a recent new species, S. pettenkoferi. S. pettenkoferi was first described in 2001 by Trülzsch et al. (16), who isolated the first two strains from a blood culture (strain B3117) and from a bursitis wound sample (strain A6667). The common trait between the two patients from whom the strains were isolated was their immunocompromised state resulting from extrapulmonary tuberculosis and lymphocytic leukemia, respectively. Our patient was also moderately immunocompromised because of his type 1 diabetes mellitus and was thus more susceptible to infection.
This case illustrates the rarity of the association of S. pettenkoferi with infection due to the inability to correctly identify this organism. Indeed, few biochemical tests are positive for this new species, which is unknown in the databases for biotypical identification. New molecular methods allow the identification of new bacteria, especially among coagulase-negative staphylococci (6). These coagulase-negative staphylococci should be considered significant when multiple deep-sample cultures are positive (8, 14). Distinguishing between contaminating and infecting isolates is a daily challenge for the microbiologist.
Footnotes
Published ahead of print on 3 January 2007.
REFERENCES
- 1.Becker, K., D. Harmsen, A. Mellmann, C. Meier, P. Schumann, G. Peters, and C. von Eiff. 2004. Development and evaluation of a quality-controlled ribosomal sequence database for 16S ribosomal DNA-based identification of Staphylococcus species. J. Clin. Microbiol. 42:4988-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Ami, R., S. Navon-Venezia, D. Schwartz, Y. Schlezinger, Y. Mekuzas, and Y. Carmeli. 2005. Erroneous reporting of coagulase-negative staphylococci as Kocuria spp. by the Vitek 2 system. J. Clin. Microbiol. 43:1448-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cunha, M. L., Y. K. Sinzato, and L. V. Silveira. 2004. Comparison of methods for the identification of coagulase-negative staphylococci. Mem. Inst. Oswaldo Cruz 99:855-860. [DOI] [PubMed] [Google Scholar]
- 4.Drancourt, M., and D. Raoult. 2002. rpoB gene sequence-based identification of Staphylococcus species. J. Clin. Microbiol. 40:1333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fontana, C., M. Favaro, M. Pelliccioni, E. S. Pistoia, and C. Favalli. 2005. Use of the MicroSeq 500 16S rRNA gene-based sequencing for identification of bacterial isolates that commercial automated systems failed to identify correctly. J. Clin. Microbiol. 43:615-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freney, J., W. E. Kloos, V. Hajek, J. A. Webster, M. Bes, Y. Brun, and C. Vernozy-Rozand. 1999. Recommended minimal standards for description of new staphylococcal species. Subcommittee on the taxonomy of staphylococci and streptococci of the International Committee on Systematic Bacteriology. Int. J. Syst. Bacteriol. 49:489-502. [DOI] [PubMed] [Google Scholar]
- 7.Ieven, M., J. Verhoeven, S. R. Pattyn, and H. Goossens. 1995. Rapid and economical method for species identification of clinically significant coagulase-negative staphylococci. J. Clin. Microbiol. 33:1060-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kloos, W. E., and T. L. Bannerman. 1994. Update on clinical significance of coagulase-negative staphylococci. Clin. Microbiol. Rev. 7:117-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martineau, F., F. J. Picard, D. Ke, S. Paradis, P. H. Roy, M. Ouellette, and M. G. Bergeron. 2001. Development of a PCR assay for identification of staphylococci at genus and species levels. J. Clin. Microbiol. 39:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mellman, A., K. Becker, C. von Eiff, U. Keckevoet, P. Schumann, and D. Harmsen. 2006. Sequencing and staphylococci identification. Emerg. Infect. Dis. 12:333-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mollet, C., M. Drancourt, and D. Raoult. 1997. rpoB sequence analysis as a novel basis for bacterial identification. Mol. Microbiol. 26:1005-1011. [DOI] [PubMed] [Google Scholar]
- 12.Poyart, C., G. Quesne, C. Boumaila, and P. Trieu-Cuot. 2001. Rapid and accurate species-level identification of coagulase-negative staphylococci by using the sodA gene as a target. J. Clin. Microbiol. 39:4296-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rantakokko-Jalava, K., S. Nikkari, J. Jalava, E. Eerola, M. Skurnik, O. Meurman, O. Ruuskanen, A. Alanen, E. Kotilainen, P. Toivanen, and P. Kotilainen. 2000. Direct amplification of rRNA genes in diagnosis of bacterial infections. J. Clin. Microbiol. 38:32-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rupp, M. E., and G. L. Archer. 1994. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin. Infect. Dis. 19:231-243. [DOI] [PubMed] [Google Scholar]
- 15.Sivadon, V., M. Rottman, S. Chaverot, J. C. Quincampoix, V. Avettand, P. de Mazancourt, L. Bernard, P. Trieu-Cuot, J. M. Feron, A. Lortat-Jacob, P. Piriou, T. Judet, and J. L. Gaillard. 2000. Use of genotypic identification by sodA sequencing in a prospective study to examine the distribution of coagulase-negative Staphylococcus species among strains recovered during septic orthopedic surgery and evaluate their significance. J. Clin. Microbiol. 43:2952-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trülzsch, K., H. Rinder, J. Trèek, L. Bader, U. Wilhelm, and J. Heesemann. 2002. “Staphylococcus pettenkoferi,” a novel staphylococcal species isolated from clinical specimens. Diagn. Microbiol. Infect. Dis. 43:175-182. [DOI] [PubMed] [Google Scholar]