Abstract
Data on clinical isolates of Kodamaea (Pichia) ohmeri, an emerging fungal pathogen, are scarce. Over the past 5 years, we identified yeast isolates from nine patients with fungemia as K. ohmeri by using the API 20C system. Here, we reanalyzed these isolates first by sequencing the internal transcribed spacer 2 (ITS2) regions and then by growing the isolates on CHROMagar Candida medium and subjecting them to pulsed-field gel electrophoresis (PFGE). Based on their ITS2 sequences, six of the nine isolates were confirmed as K. ohmeri, while the others were identified as Candida haemulonii (n = 2) and Candida parapsilosis (n = 1). PFGE karyotyping of the K. ohmeri isolates revealed similar major bands, and their colonies showed a characteristic color change from pink to blue when grown on CHROMagar Candida medium for more than 48 h. For K. ohmeri, the ranges of MICs of fluconazole, voriconazole, caspofungin, and micafungin were 2 to 32 μg/ml, 0.03 to 0.5 μg/ml, 0.125 to 0.25 μg/ml, and 0.03 to 0.06 μg/ml, respectively. Restriction endonuclease analysis of genomic NotI-digested DNA (REAG-N) from isolates from different patients produced unique patterns, suggesting that the fungemia had occurred sporadically. This study determined that ITS2 sequence data, PFGE karyotypes, and CHROMagar Candida chromogenic culture medium are reliable diagnostic tools for identifying K. ohmeri while REAG-N is useful for genotyping the clinical isolates of K. ohmeri.
Concern regarding systemic infections caused by unusual fungi has grown as the global population of immunocompromised patients has increased. Some rare yeast species may be inherently resistant to antifungal agents (8), leading to the development of nosocomial clusters; therefore, rapid and accurate identification is essential for the proper treatment and management of infections. Kodamaea (Pichia) ohmeri, an ascosporogenous yeast and a teleomorph of Candida guilliermondii var. membranaefaciens, is an environmental strain commonly used in the food industry for the fermentation of pickles, rinds, and fruit; however, it is also an emerging fungal pathogen, particularly in immunocompromised patients (3, 7). To date, 12 cases of K. ohmeri infection have been reported, including nine cases presenting with fungemia plus two cases that occurred as a nosocomial cluster (3, 7, 12). Although molecular biological techniques that allow more precise identification and genotyping of yeast species have recently been developed, little research has been conducted on identifying and genotyping the clinical isolates of K. ohmeri.
Over the past 5 years, we identified yeast isolates from nine patients as K. ohmeri by using the API 20C assimilation test (bioMérieux, Marcy L'Etoile, France) at the Clinical Microbiology Laboratory in Chonnam National University Hospital, Gwangju, Korea. We had limited confidence in the accuracy of this identification, however, because the performance of commercial identification systems with rare and unusual yeasts varies considerably (16). We therefore sought to precisely identify and genotype each of the previously tested isolates by sequencing the internal transcribed spacer 2 (ITS2) region of the rRNA gene followed by pulsed-field gel electrophoresis (PFGE). We also used the API 20C and Vitek 2 yeast card (YST) systems (bioMérieux), as well as CHROMagar Candida chromogenic growth medium, to identify each isolate. Finally, we tested the susceptibility of K. ohmeri to select antifungal agents.
MATERIALS AND METHODS
Isolates and conventional identification.
We analyzed 16 yeast isolates (13 bloodstream isolates, 2 isolates from catheter sites, and 1 isolate from a phlebitis site) from nine patients with fungemia; the isolates were previously identified as K. ohmeri by the traditional identification methods based on API 20C. Fourteen isolates from seven patients (patients 1 to 6 and patient 9) were obtained from clinical specimens as part of routine diagnostic procedures performed at Chonnam National University Hospital (a 1,000-bed tertiary-care hospital in Gwangju, Korea) from January 1999 to December 2003; two isolates from two patients (patients 7 and 8) were referred by the Asan Medical Center (a 2,200-bed tertiary-care hospital in Seoul, Korea) for the identification. We used K. ohmeri ATCC 46051 as a control in this study. All K. ohmeri isolates were reidentified via assessment of API 20C sugar assimilation patterns and the use of the Vitek 2 system (Vitek 2 ID-YST) and CHROMagar Candida medium (BBL, Beckton Dickinson, Sparks, MD).
Amplification and sequencing of the ITS2 region.
The ITS2 region, which is located between the 5.8S and 28S subunits in the rRNA gene, was sequenced for accurate identification of the isolates (1, 5). The first isolates collected from each of the nine patients were analyzed. DNA from the isolates was extracted by previously described methods (4). The fungus-specific universal primers ITS3 (5′-GCATCGATGAAGAACGCAGC-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) were used to amplify the ITS2 region (19). All loci were sequenced in both the forward and reverse directions with the same primers as those used for the PCR. The PCR was performed with a total reaction mixture volume of 50 μl consisting of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 1.2 U of GoTaq DNA polymerase (Promega Corporation, Madison, WI), 0.4 μM (each) ITS2 region primers (ITS3/ITS4), and 2 μl (1 to 5 ng) of DNA template. PCR was carried out using the following conditions: initial denaturation at 94°C for 5 min; 30 cycles of denaturation (94°C for 30 s), annealing (55°C for 30 s), and extension (72°C for 30 s); and a final extension step at 72°C for 5 min. The PCR products were purified and sequenced using the ABI 3730XL sequencer (Applied Biosystems, Foster City, CA). Sequence data were assembled and compared with previously reported sequences from two K. ohmeri strains (GenBank accession no. AY382339 and AF218977) by using DNA Sequencher software (Gene Codes Corp., Ann Arbor, MI).
PFGE analysis.
The PFGE analyses were conducted according to a previously described procedure (13-15). PFGE typing consisted of electrophoretic karyotyping (EK) and restriction endonuclease analysis of genomic DNA by using NotI (REAG-N). In brief, one colony of each yeast isolate from the 48-h Sabouraud dextrose agar (SDA) cultures was incubated overnight at 37°C in 10 ml of YPD broth (glucose, 2%; yeast extract, 1%; Bacto-peptone, 2% [Difco]). A 150-μl aliquot of the cell suspension was mixed evenly with 30 U of lyticase (Sigma, St. Louis, MO) and 150 μl of 1.6% low-melting-temperature agarose (FMC BioProducts, Hercules, CA), which was previously melted, and kept liquid at 50 to 55°C. Aliquots placed in plug molds were incubated at room temperature for 20 min. The agarose plugs were removed from the plug molds and placed in 500 μl of a lyticase buffer containing 50 mM EDTA and 100 U of lyticase/ml for 2 h and then washed once in 2 ml of distilled water. The plugs were incubated in proteinase K solution (50 mM EDTA and 100 μg of proteinase K; Invitrogen, Carlsbad, CA) for 16 to 18 h at 50°C and finally washed five times in 50 mM sodium-EDTA (pH 8.0). Yeast chromosomal DNA was separated by PFGE using the GenePath system (Bio-Rad, Hercules, CA). Electrophoresis was performed for 48 h in 0.8% agarose gel (SeaKem GTG agarose; FMC BioProducts) in 0.5× TBE buffer (0.1 M Tris, 0.09 M boric acid, 0.01 M EDTA, pH 8.0) at 4 V/cm with initial and final switch times of 90 and 325 s, respectively. Immediately following the electrophoresis, the gels were stained with 0.5 μg/ml ethidium bromide solution and then photographed under UV illumination. Isolates that were determined to differ by one or more bands were designated as possessing different karyotypes (14, 15, 17).
For REAG, digestion was carried out with NotI at 37°C for 16 h. Electrophoresis for REAG with NotI was performed using the EK method, except that 1.2% agarose gel (SeaKem GTG agarose; FMC BioProducts) was used. Electrophoresis for REAG with NotI was performed for 40 h in 1.2% agarose gel in 0.5× TBE buffer (0.1 M Tris, 0.09 M boric acid, 0.01 M EDTA, pH 8.0) at 4 V/cm with initial and final switch times of 5.3 and 49.9 s, respectively. Isolates were considered identical or similar when ≥95% of the bands matched. Isolates with less than 95% of bands matching were considered different (18). All of the isolates in this study were analyzed at least twice, which involved the subculturing of isolates from the original stock culture to Sabouraud dextrose agar, DNA preparation, endonuclease digestion, and the separation of the DNA via PFGE in order to discern pattern relationships and ensure reproducibility.
Antifungal susceptibility testing.
Broth microdilution testing was performed in accordance with the guidelines of the Clinical and Laboratory Standards Institute (formerly NCCLS) document M27-A2 (2) by using RPMI 1640 medium and an inoculum of 0.5 × 103 to 2.5 × 103 cells/ml. The final concentrations of the antifungal agents were 0.313 to 16 μg/ml for amphotericin B (Sigma, St. Louis, MO), itraconazole (Janssen Pharmaceutica, Beerse, Belgium), voriconazole (Pfizer, Sandwich, United Kingdom), caspofungin (Merck, Whitehouse Station, NJ), and micafungin (Astellas Pharma Inc., Tokyo, Japan) and 0.125 to 64 μg/ml for fluconazole (Pfizer). The trays were incubated in air at 35°C, and MIC end points were read after 24 h for caspofungin and micafungin (6, 9) and after 48 h of incubation for all other drugs (2). The MIC of amphotericin B was defined as the lowest concentration resulting in the complete inhibition of growth, while the MICs of fluconazole, itraconazole, voriconazole, caspofungin, and micafungin were defined as the lowest concentrations that produced a prominent decrease in turbidity (approximately 50%) relative to a drug-free control well (2, 6, 9). Two reference strains, Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258, were tested as quality control isolates for each antifungal susceptibility test.
Clinical data analysis.
The charts of the fungemia patients whose blood isolates were finally identified as K. ohmeri were reviewed retrospectively. The demographic and clinical data included gender, age, underlying conditions, dates of positive blood cultures, dates of antifungal drug administration and drug doses, information on catheter removal and culturing, and the outcome of the fungemia (13, 15). Neutropenia was defined as a neutrophil count of fewer than 1,000/mm3 at the time of the onset of infection.
RESULTS AND DISCUSSION
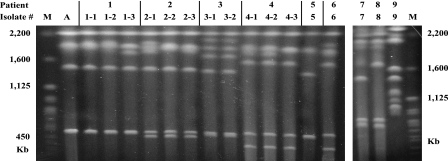
The identification of medically important yeasts by ITS sequencing, especially sequencing of the ITS2 region, is a reliable and accurate alternative to conventional identification methods (5). In fact, three reference strains of K. ohmeri not identified by ITS1 sequencing were accurately identified by their ITS2 sequences, indicating that the sequence of ITS2 may be more species-specific than that of ITS1 (5). In this study we used ITS2 sequencing to analyze yeast isolates, collected from nine patients, that had previously been identified as K. ohmeri by using the API 20C system. The first isolates collected from six of the nine patients were identified as K. ohmeri, showing 100.0% homology with the published K. ohmeri ITS2 sequence (GenBank accession no. AY382339 or AF218977). The remaining three first-collected isolates were identified as Candida haemulonii (n = 2) and C. parapsilosis (n = 1) (Table 1). Among the six isolates that were identified as K. ohmeri by ITS2 sequencing, all of the strains contained three to six chromosomes that ranged in size from 300 to 2,000 kb (Fig. 1). Similarly, electrophoretic karyotyping (EK) produced two bands (500 and 1,900 kb) not only for all 13 K. ohmeri isolates from six patients but also for a reference strain (K. ohmeri ATCC 46051), in accordance with previous findings (11). These data suggest that K. ohmeri species identification can be accomplished by comparing the EK patterns of patient strains with that of a reference strain. Overall, isolates from six of the nine patients were confirmed as K. ohmeri by using both ITS2 sequencing and EK analysis.
TABLE 1.
Results of identification testing for nine yeast strains in this study
| Patient | Isolate no. | Identification by ITS2 sequence analysisa | API 20C
|
Vitek 2 ID-YST resultc | Color change (pink to blue) of colonies on CHROMagar | |
|---|---|---|---|---|---|---|
| Resultb | Code no. | |||||
| 1 | 1-1 | K. ohmeri (100) | K. ohmeri (76.2) | 7356373 | K. ohmeri (excellent) | Yes |
| 2 | 2-1 | K. ohmeri (100) | K. ohmeri (99.9) | 7156376 | K. ohmeri (low) | Yes |
| 3 | 3-1 | K. ohmeri (100) | K. ohmeri (99.9) | 7376777 | K. ohmeri (low) | Yes |
| 4 | 4-1 | K. ohmeri (100) | K. ohmeri (99.7) | 6356376 | K. ohmeri (excellent) | Yes |
| 5 | 5-1 | K. ohmeri (100) | K. ohmeri (56.1) | 6352376 | K. ohmeri (excellent) | Yes |
| 6 | 6-1 | K. ohmeri (100) | K. ohmeri (99.7) | 7356377 | K. ohmeri (excellent) | Yes |
| 7 | 7-1 | C. haemulonii | K. ohmeri (99.9) | 6142176 | No identification | No (lavender) |
| 8 | 8-1 | C. haemulonii | K. ohmeri (99.9) | 6142176 | No identification | No (lavender) |
| 9 | 9-1 | C. parapsilosis | K. ohmeri (88.1) | 6146177 | C. intermedia (low) | No (lavender) |
Numbers in parentheses are percentages of homology of the ITS2 sequences of the isolates to the reference sequence of K. ohmeri.
Numbers in parentheses are probabilities of correct identification (%).
The probability of correct identification is indicated in parentheses.
FIG. 1.
EK of yeast isolates identified as K. ohmeri by using conventional methods (Tables 1 and 2). All strains from six patients (patients 1 to 6) whose isolates were identified as K. ohmeri by ITS2 sequencing and a reference strain (isolate A) had similar major EK bands, suggesting that EK is effective for distinguishing K. ohmeri from other species. Serial isolates from patients 1, 2, and 4 showed one or two differences in banding at sizes greater than 1,600 kb. A, K. ohmeri ATCC 46051. M, Saccharomyces cerevisiae DNA concatemers as molecular size markers.
Although API 20C is considered to be the “gold standard” for the identification of yeast species, our study showed that three Candida strains (two strains of C. haemulonii and one strain of C. parapsilosis) were misidentified as K. ohmeri by using the API 20C system. In addition, although the Vitek 2 ID-YST system correctly identified the isolates from the six patients as K. ohmeri, two (33%) of the six first-collected isolates were identified with low discrimination. Similarly, Rodero et al. (10) reported that the Vitek system misidentified a C. haemulonii isolate as K. ohmeri (86% probability of identity).
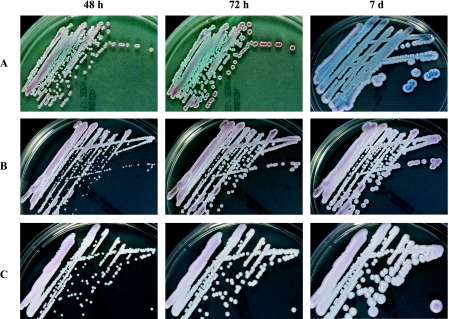
CHROMagar Candida chromogenic growth medium is an extremely useful tool to assist in making an identification of Candida species based on the development of colored colonies. We found that the colonies of all 13 K. ohmeri isolates from the six patients, in addition to that of K. ohmeri strain ATCC 46051, underwent a characteristic change from pink to blue over a 48-h period (Fig. 2). Otag et al. (7) also reported pink-blue colonies for the morphotype of K. ohmeri on CHROMagar media. Our laboratory has used CHROMagar Candida growth medium for over 10 years, and in our experience, few other yeast species share the color change characteristic of K. ohmeri on CHROMagar plates. Thus, CHROMagar Candida chromogenic growth medium is a simple and useful tool for the identification of K. ohmeri. In clinical microbiology laboratories, culture on CHROMagar Candida medium and biochemical investigations via either the API or Vitek system can be routinely applied to accurately identify K. ohmeri isolates from clinical specimens; however, at least 2 to 3 days are required to obtain pink-blue colonies on CHROMagar plates and a full week is required for K. ohmeri to complete its blue color development. Thus, faster molecular diagnostic tools for the proper identification of fungal pathogens should also be evaluated.
FIG. 2.
Macroscopic morphology of K. ohmeri (A) grown at 35°C on CHROMagar Candida medium for 48 h, 72 h, and 1 week. Note the characteristic color change of K. ohmeri from pink to blue, in contrast to the control fungal isolates (C. haemulonii [B] and C. parapsilosis [C]).
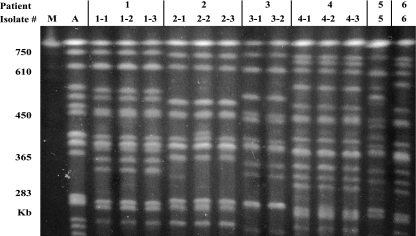
The molecular typing of K. ohmeri by PFGE has not previously been reported. In this study, 13 K. ohmeri isolates from six patients with fungemia were analyzed by PFGE, revealing six different REAG-N types (Table 2 and Fig. 2). Additionally, for four patients with serial bloodstream isolates and serial isolates from other sites, all of the identified strains from each patient had the same REAG-N patterns (Fig. 3). The K. ohmeri isolates from the six different patients showed unique REAG-N patterns, suggesting that the fungemia had occurred sporadically; however, EK revealed that serial isolates from the same patients fell into two or three different EK types for three of four patients, revealing 10 different karyotypes for 13 isolates from six patients (Fig. 1). Minor (one- or two-band) differences in the karyotypes, which may have been due to chromosomal instability or rearrangements within a single strain, were detected only at sizes of >1,600 kb. Because yeast isolates that differ by one or more bands are generally considered to have different karyotypes (14, 15, 17), EK may be of limited value for the epidemiological typing of K. ohmeri isolates compared to REAG-N. When we compared isolates obtained from catheters (patients 2 and 4) or at a phlebitis site (patient 1) with blood isolates from the same individuals, the K. ohmeri isolates recovered from each patient had identical REAG-N patterns regardless of the collection site. Our findings suggest that the organism entered these patients transvenously, via a catheter, and that REAG-N analysis is very effective for the epidemiological typing of clinical strains of K. ohmeri.
TABLE 2.
Genotyping and antifungal susceptibility testing results for K. ohmeri isolates from blood and various body sites of six patients
| Patient | Isolate no. | Source | Isolation date (mo/day/yr) | PFGE pattern designation
|
MICa (μg/ml) of:
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| EK | REAG-N | AmB | Flu | Itra | Vori | Casp | Mica | ||||
| 1 | 1-1 | Blood | 08/30/1999 | K1 | R1 | 0.5 | 4 | 0.125 | 0.06 | 0.25 | 0.06 |
| 1-2 | Blood | 09/12/1999 | K1 | R1 | 0.5 | 4 | 0.125 | 0.06 | 0.25 | 0.06 | |
| 1-3 | Phlebitis siteb | 09/05/1999 | K2 | R1 | 0.5 | 4 | 0.125 | 0.06 | 0.125 | 0.06 | |
| 2 | 2-1 | Blood | 07/31/2000 | K3 | R2 | 0.25 | 32 | 0.5 | 0.5 | 0.125 | 0.03 |
| 2-2 | Catheter | 08/09/2000 | K4 | R2 | 0.25 | 32 | 0.5 | 0.5 | 0.125 | 0.03 | |
| 2-3 | Blood | 08/14/2000 | K5 | R2 | 0.5 | 32 | 0.5 | 0.5 | 0.125 | 0.03 | |
| 3 | 3-1 | Blood | 11/14/2002 | K6 | R3 | 0.25 | 16 | 0.25 | 0.125 | 0.25 | 0.06 |
| 3-2 | Blood | 11/21/2002 | K6 | R3 | 0.25 | 16 | 0.25 | 0.125 | 0.25 | 0.06 | |
| 4 | 4-1 | Blood | 11/15/2003 | K7 | R4 | 0.25 | 4 | 0.125 | 0.03 | 0.125 | 0.03 |
| 4-2 | Catheter | 11/13/2003 | K7 | R4 | 0.25 | 4 | 0.125 | 0.03 | 0.125 | 0.03 | |
| 4-3 | Blood | 11/04/2003 | K8 | R4 | 0.5 | 4 | 0.125 | 0.03 | 0.125 | 0.03 | |
| 5 | 5-1 | Blood | 08/01/2000 | K9 | R5 | 0.25 | 16 | 0.125 | 0.06 | 0.125 | 0.03 |
| 6 | 6-1 | Blood | 07/03/2001 | K10 | R6 | 0.5 | 2 | 0.125 | 0.03 | 0.125 | 0.03 |
AmB, amphotericin B; Flu, fluconazole; Itra, itraconazole; Vori, voriconazole; Casp, caspofungin; Mica, micafungin.
A swab sample from the skin surrounding the inflamed vein (which had already been removed via a peripheral venous catheter) in the right leg.
FIG. 3.
REAG-N followed by PFGE for K. ohmeri isolates obtained from six patients. Among the 13 isolates obtained from six patients, six different types were identified by REAG-N, and serial isolates from the same patients (patients 1 to 4) had the same REAG-N patterns. The K. ohmeri isolates from the six different patients showed unique REAG-N patterns, suggesting that the fungemia had occurred sporadically. A, K. ohmeri ATCC 46051. M, Saccharomyces cerevisiae DNA concatemers as molecular size markers.
We also tested the susceptibility of K. ohmeri to various antifungal agents. Similar to findings in previous reports (3, 7), in this study all of the isolates of K. ohmeri were susceptible to amphotericin. In addition, the isolates demonstrated low susceptibility to fluconazole and itraconazole, with some isolates showing a dose-dependent response (Table 2). To date, few data exist regarding the susceptibility of K. ohmeri to the antifungals voriconazole and echinocandin. In our study, the voriconazole MICs for the isolates ranged from 0.03 to 0.5 μg/ml, suggesting that voriconazole may be active against strains of K. ohmeri. The ranges of MICs of caspofungin and micafungin were 0.125 to 0.25 μg/ml and 0.03 to 0.06 μg/ml, respectively. These data suggest that K. ohmeri is less susceptible to caspofungin than three common isolates of Candida known to cause bloodstream infections: Candida albicans, Candida glabrata, and Candida tropicalis are all extremely susceptible to caspofungin (MIC at which 90% of the isolates tested are inhibited, 0.06 μg/ml) (9).
Nine cases of K. ohmeri fungemia have been previously reported, including one case of K. ohmeri fungemia-associated phlebitis (in one of the patients included in our study) (12). Here, we report an additional five cases of K. ohmeri-related fungemia identified at our hospital over a 5-year period (Table 3). All of the patients had preexisting conditions and were receiving broad-spectrum antibiotics and parenteral nutrition. In addition, all five patients had central venous catheters when the first positive cultures were obtained. In two of the patients (patients 3 and 6), the fungemia cleared after catheter removal (without antifungal therapy), but the other three patients received antifungal therapy. Patient 4 was responsive to amphotericin B therapy, and this patient's death was not attributed to fungemia. In contrast, two patients (patients 2 and 5) died of fungemia despite antifungal therapy (fluconazole for patient 2 and combined amphotericin B and fluconazole therapy for patient 5). For patients 2 and 5, positive blood cultures were obtained 15 and 4 days, respectively, before each patient died. Both patients developed polymicrobial (K. ohmeri and C. krusei) fungemia and received immunosuppressive therapy. These results suggest that while most K. ohmeri infections are responsive to catheter removal and antifungal treatment, immunosuppressive therapy with combined C. krusei-related fungemia may be associated with a fatal outcome.
TABLE 3.
Clinical characteristics and therapy regimens and outcomes for six patients with K. ohmeri fungemia
| Patient | Age (yr) | Sexb | Underlying condition(s) | No. of blood cultures (days)c positive for K. ohmeri | Clinical status at time of first positive culture
|
Antifungal therapy (duration, days)c | Day of catheter removalc (result of tip culture) | Outcome (day of death)c | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Immunosuppressive state (cause) | Neutropeniad | Vascular catheter status | Treatment with broad-spectrum antibiotics | Parenteral nutrition | Previous operation | ||||||||
| 1a | 59 | M | Ventriculoperitoneal shunt infection, pneumonia | 4 (0, 6, 8, 13) | No | No | Peripheral catheter already removed | Yes | Yes | Yes | Amphotericin B, 50 mg (16 to 30) | Recovered | |
| 2 | 11 | M | Burkitt's lymphoma, C. krusei fungemia | 4 (0, 12, 13, 14) | Yes (chemotherapy) | Yes | Central venous catheter | Yes | Yes | No | Fluconazole, 50 mg (−10 to 15)e | 9 (>15 CFU) | Exitus (15) |
| 3 | 41 | M | Alcoholic ketoacidosis, tuberculosis | 3 (0, 5, 7) | No | No | Central venous catheter | Yes | Yes | Yes | None | 8 (>15 CFU) | Recovered |
| 4 | 47 | M | Pneumonia, diabetes, chronic renal failure | 10 (0, 1, 2, 3, 4, 5, 8, 9, 11, 12) | No | No | Central venous catheter | Yes | Yes | No | Amphotericin B, 200 mg (9 to 26); fluconazole, 400 mg (8) | 12 (>15 CFU) | Exitus (67) |
| 5 | 4 | F | Tetralogy of Fallot, C. krusei fungemia | 1 | Yes (immunotherapy) | No | Central venous catheter | Yes | Yes | No | Amphotericn B, 3 mg (−1 to 4); fluconazole, 8 mg (−2 to 4)e | Exitus (4) | |
| 6 | 0 | F | Prematurity with very low birth wt | 1 | Yes (prematurity) | No | Umbilical artery and vein catheter | Yes | Yes | No | None | 1 (>15 CFU) | Recovered |
Patient 1 was previously described in reference 12.
M, male; F, female.
Days are numbered relative to the time of the first positive blood culture.
Neutropenia is defined as <109 neutrophils/liter of blood.
Fungemia occurred during antifungal therapy (for the treatment of C. krusei candidemia) in patients 2 and 5.
Over the course of infection, no significant increase in the MICs of any of the six antifungal agents for the serial isolates from each of four patients with fungemia was observed. However, when the MICs for the isolates from different patients were compared, the isolates from patient 2 were found to be approximately three- or fourfold less susceptible to all three azole antifungals than the isolates from the other patients. Although a low dose of fluconazole was administered (50 mg for 10 days) prior to the first positive blood culture, it is unknown whether the K. ohmeri strains in this patient were inherently or secondarily resistant to azoles. Patient 2 was treated with fluconazole alone and did not survive, suggesting that amphotericin B or other antifungal therapy would have been appropriate.
Due to the increased incidence of infectious diseases caused by less common yeast species, most laboratories have instituted routine testing for yeast identification, including such commercially available reagents as the API 20C and Vitek systems and CHROMagar Candida growth medium. Because K. ohmeri has low susceptibility to azole antifungal agents and may cause nosocomial clusters, rapid and accurate identification is essential. Our study demonstrates that color change on CHROMagar Candida medium is a simple and reliable diagnostic tool for identifying K. ohmeri. In addition, molecular analysis is useful for species identification (via ITS2 sequencing and EK) and epidemiological typing (via REAG-N) of bloodstream strains of K. ohmeri.
Footnotes
Published ahead of print on 24 January 2007.
REFERENCES
- 1.Chen, Y. C., J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barrett, K. LaFe, S. L. Yarfitz, A. P. Limaye, and B. T. Cookson. 2000. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J. Clin. Microbiol. 38:2302-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard M27-A2, 2nd ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.Han, X. Y., J. J. Tarrand, and E. Escudero. 2004. Infections by the yeast Kodomaea (Pichia) ohmeri: two cases and literature review. Eur. J. Clin. Microbiol. Infect. Dis. 23:127-130. [DOI] [PubMed] [Google Scholar]
- 4.Hoffman, C. S. 1993. Preparation of yeast DNA, p. 13.11.1-13.11.4. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley and Sons, New York, NY.
- 5.Leaw, S. N., H. C. Chang, H. F. Sun, R. Barton, J. P. Bouchara, and T. C. Chang. 2006. Identification of medically important yeast species by sequence analysis of the internal transcribed spacer regions. J. Clin. Microbiol. 44:693-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Odds, F. C., M. Motyl, R. Andrade, J. Bille, E. Canton, M. Cuenca-Estrella, A. Davidson, C. Durussel, D. Ellis, E. Foraker, A. W. Fothergill, M. A. Ghannoum, R. A. Giacobbe, M. Gobernado, R. Handke, M. Laverdiere, W. Lee-Yang, W. G. Merz, L. Ostrosky-Zeichner, J. Peman, S. Perea, J. R. Perfect, M. A. Pfaller, L. Proia, J. H. Rex, M. G. Rinaldi, J. L. Rodriguez-Tudela, W. A. Schell, C. Shields, D. A. Sutton, P. E. Verweij, and D. W. Warnock. 2004. Interlaboratory comparison of results of susceptibility testing with caspofungin against Candida and Aspergillus species. J. Clin. Microbiol. 42:3475-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otag, F., N. Kuyucu, Z. Erturan, S. Sen, G. Emekdas, and T. Sugita. 2005. An outbreak of Pichia ohmeri infection in the paediatric intensive care unit: case reports and review of the literature. Mycoses 48:265-269. [DOI] [PubMed] [Google Scholar]
- 8.Pfaller, M. A., and D. J. Diekema. 2004. Rare and emerging opportunistic fungal pathogens: concern for resistance beyond Candida albicans and Aspergillus fumigatus. J. Clin. Microbiol. 42:4419-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfaller, M. A., S. A. Messer, L. Boyken, C. Rice, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2004. Further standardization of broth microdilution methodology for in vitro susceptibility testing of caspofungin against Candida species by use of an international collection of more than 3,000 clinical isolates. J. Clin. Microbiol. 42:3117-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodero, L., M. Cuenca-Estrella, S. Cordoba, P. Cahn, G. Davel, S. Kaufman, L. Guelfand, and J. L. Rodriguez-Tudela. 2002. Transient fungemia caused by an amphotericin B-resistant isolate of Candida haemulonii. J. Clin. Microbiol. 40:2266-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosa, C. A., M. A. Lachance, W. T. Starmer, J. S. Barker, J. M. Bowles, and B. Schlag-Edler. 1999. Kodamaea nitidulidarum, Candida restingae and Kodamaea anthophila, three new related yeast species from ephemeral flowers. Int. J. Syst. Bacteriol. 49:309-318. [DOI] [PubMed] [Google Scholar]
- 12.Shin, D. H., J. H. Park, J. H. Shin, S. P. Suh, D. W. Ryang, and S. J. Kim. 2003. Pichia ohmeri fungemia associated with phlebitis: successful treatment with amphotericin B. J. Infect. Chemother. 9:88-89. [DOI] [PubMed] [Google Scholar]
- 13.Shin, J. H., M. N. Kim, D. H. Shin, S. I. Jung, K. J. Kim, D. Cho, S. J. Kee, M. G. Shin, S. P. Suh, and D. W. Ryang. 2004. Genetic relatedness among Candida tropicalis isolates from sporadic cases of fungemia in two university hospitals in Korea. Infect. Control Hosp. Epidemiol. 25:634-640. [DOI] [PubMed] [Google Scholar]
- 14.Shin, J. H., M. R. Park, J. W. Song, D. H. Shin, S. I. Jung, D. Cho, S. J. Kee, M. G. Shin, S. P. Suh, and D. W. Ryang. 2004. Microevolution of Candida albicans strains during catheter-related candidemia. J. Clin. Microbiol. 42:4025-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin, J. H., D. H. Shin, J. W. Song, S. J. Kee, S. P. Suh, and D. W. Ryang. 2001. Electrophoretic karyotype analysis of sequential Candida parapsilosis isolates from patients with persistent or recurrent fungemia. J. Clin. Microbiol. 39:1258-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verweij, P. E., I. M. Breuker, A. J. Rijs, and J. F. Meis. 1999. Comparative study of seven commercial yeast identification systems. J. Clin. Pathol. 52:271-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voss, A., R. J. Hollis, M. A. Pfaller, R. P. Wenzel, and B. N. Doebbeling. 1994. Investigation of the sequence of colonization and candidemia in nonneutropenic patients. J. Clin. Microbiol. 32:975-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voss, A., M. A. Pfaller, R. J. Hollis, J. Rhine-Chalberg, and B. N. Doebbeling. 1995. Investigation of Candida albicans transmission in a surgical intensive care unit cluster by using genomic DNA typing methods. J. Clin. Microbiol. 33:576-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA.