Abstract
Fungal infections due to Candida species represent an important cause of nosocomial bloodstream infections. We report a large pseudo-outbreak of Candida guilliermondii fungemia that occurred in a university hospital in Brazil. C. guilliermondii was identified in 64 (43%) of the 149 blood samples drawn between June 2003 and July 2004. The samples were from patients in different wards of the hospital but concentrated in pediatric units. None of the patients had clinical signs of fungemia, and observational analysis revealed errors in the collection of blood samples. During the investigation of the pseudo-outbreak, C. guilliermondii was isolated from environmental surfaces and from the skin and nails of members of the nursing team. Through a subtyping analysis it was found that some of the nonpatient isolates were highly related to the patient isolates, and all the patient isolates were highly related. This is consistent with the hypothesis that the pseudo-outbreak was from a limited number of common sources. The adoption of intervention measures was effective in resolving the outbreak, supporting the hypothesis that the outbreak was due to poor techniques of drawing blood samples for culture.
Fungal infections due to Candida species represent an important cause of nosocomial bloodstream infections (18, 20). Candida is the fourth most prevalent cause of nosocomial bloodstream infections in Brazil, with an incidence of 2.49 cases per 1,000 hospital admissions, and a crude mortality of 54% (1). Candida guilliermondii is an infrequent, but not unusual, agent of candidiasis and has been described as an emerging pathogen. As reviewed by Krcmery and Barnes (7), over a 40-year period ending in 1990, only 10 cases of C. guilliermondii fungemia had been reported, whereas 33 cases were reported between 1990 and 2002. In that study, the rate of fungemia due to C. guilliermondii varied from 0 to 0.7% in the period between 1950 and 1990 and from 0.7 to 5.5% from 1991 to 1998. Most cases of C. guilliermondii infection are associated with oncology patients (8, 16, 19). A review of the cases of fungemia in oncology patients reported from 1960 to 1990 showed that 10 cases (0.7%) were due to C. guilliermondii, with three deaths (19). In Brazil, a study conducted in six hospitals from São Paulo and Rio de Janeiro by Colombo et al. (2) showed three cases of C. guilliermondii infection among 145 Candida bloodstream infection patients. In another study involving 11 Brazilian institutions, C. guilliermondii accounted for 2.4% of 712 candidemias (1). Outbreak investigations involving these organisms must distinguish reliably among the various species involved. Most strains of C. guilliermondii are morphologically and biochemically indistinguishable from Candida famata (teleomorph Debaromyces hansenii), and nucleic-acid based methods are usually required to distinguish reliably between the two species. Recently, C. guilliermondii has been further subdivided by the addition of two new species in the C. guilliermondii clade: Pichia caribbica (Candida fermentati) and Candida carpophila (15). These three species are also biochemically indistinguishable. The objective of the present study was to investigate a large pseudo-outbreak due to C. guilliermondii isolated from blood cultures during the period between April 2003 and July 2004.
MATERIALS AND METHODS
Background.
Hospital São Paulo is a teaching hospital associated with the Federal University of São Paulo (UNIFESP), located in São Paulo, Brazil. It has around 680 beds and several units, some of them for pediatric patients, distributed in 10 wards, and one emergency department.
Observational analysis.
The isolation of a yeast-like fungus identified as C. guilliermondii in blood cultures mainly obtained from the Pediatric Emergency Department (Pediatric ED) of Hospital São Paulo was noticed during epidemiological surveillance in July 2003. C. guilliermondii was isolated from patients without previous exposure to risk factors for candidemia and who did not present any clinical evidence of systemic infection. After a retrospective analysis of the data relative to the isolation of C. guilliermondii, an investigation was started by the Hospital Infection Control Committee (HICC).
Considering the possibility of a pseudo-outbreak, an observational study was performed by the epidemiologist, clinicians, and nurses of the HICC to evaluate the appropriateness of the techniques used for drawing blood samples for culture at the pediatric ED. The collection procedures were observed for 2 hours every shift for 10 days in January 2003. Individuals collected data on drawing blood, including hand hygiene procedures, use of individual protection devices, and preparation of the patient's skin. There was no intervention from the observers.
We evaluated the adherence to the local guidelines for blood sample collection (see Table 3). The local guidelines are as follows. Place a tourniquet above the venipuncture site. Palpate and locate the vein. Decontaminate skin. It is critical to disinfect the venipuncture site meticulously with 10% povidone iodine or 70% isopropyl alcohol by swabbing the skin concentrically from the center of the venipuncture site outwards. Let the disinfectant evaporate. Do not repalpate the vein again. Perform venipuncture. Decontaminate the rubber septum of the blood vial with 70% alcohol only. Obtain a 20-ml sample and divide it between aerobic and anaerobic blood culture bottles. It is recommended that two sets be collected in a 24-hour period. Specimens should not be drawn through the catheter or cannula unless a concomitant peripheral draw is obtained. The blood culture bottles should be sent immediately to the microbiology laboratory.
TABLE 3.
Parameters considered during the observational analysis of the collection of blood samples and resulting observations (n = 15)
| Parameter | No. of observations (%) |
|---|---|
| Product for hand hygiene | |
| Common soap | 11 (73.3) |
| Chlorhexidine (2%) | 3 (20.0) |
| None | 1 (6.7) |
| Product for aseptic prepn of the puncture site | |
| Common soap | 9 (60.0) |
| Iodopolvidine solution | 1 (6.7) |
| 70% alcohol | 5 (33.3) |
| Type of gloves | |
| Nonsterile (procedure) gloves | 7 (46.6) |
| Sterile gloves | 8 (53.4) |
Collection and processing of samples.
In the pediatric unit, multiple environmental cultures for fungi were obtained from povidone iodine and alcohol antiseptic solution, paper towels, and cotton. Each selected material was placed in a flask with 250 ml sterile distilled water and allowed to settle. The total volume was centrifuged at 4,000 rpm for 15 min, the supernatant was removed, and the pellet was streaked on plates containing Sabouraud dextrose agar with chloramphenicol.
A wet cotton swab was used for collecting samples from a sphygmomanometer cuff, soap tray, tray, sink, and drain. Each swab was streaked on the surface of a plate containing Sabouraud dextrose agar with chloramphenicol.
All 32 members of the nursing team of the pediatric unit were examined for evidence of dermatological disease. Samples from toenails and fingernails were collected with a sterile scalpel, and the resulting fragments were checked for fungal elements by direct microscopy as well as cultured on plates containing Sabouraud dextrose agar with chloramphenicol. Hand colonization was checked by sampling the palms and fingers of all nurses with a sterile filter paper containing 0.02% Tween 80. The collected filter papers were placed in 50-ml Falcon tubes and shaken for 15 min. The liquid was removed and centrifuged at 4,000 rpm for 15 min. The supernatant was discarded, and the pellet was streaked on plates containing Sabouraud dextrose agar with chloramphenicol.
All cultures were incubated at 30°C for 5 days. Isolated colonies were subcultured on chromogenic medium CHROMagar Candida (CHROMagar Microbiology, Paris, France) and incubated at 30°C for 72 h. Colonies representative of the cultures were individually picked for identification. Species identification was based on characteristics of micromorphology, and biochemical tests were performed with the commercial system ID 32 C (bioMérieux, Marcy l'Étoile, France).
DNA subtyping.
A total of 54 isolates of C. guilliermondii, C. famata, and related species were used for the subtyping analysis (Table 1). These isolates were classified into three groups. Group 1 consisted of 25 isolates from the hospital cluster. These are termed Brazilian cluster (or outbreak) strains. Twenty of these isolates were from blood cultures. The remaining five isolates, labeled 21 to 25, were from three nurses' hands, and two environmental sources related to the cluster (sphygmomanometer cuff and paper towel). Group 2 consisted of nine isolates from South America that were not involved in the cluster and were obtained as a kind gift from M. Pfaller (University of Iowa, Iowa City). They are termed South American control strains. Group 3 consisted of 20 unrelated strains from the CDC reference collection. These are termed U.S. nonoutbreak control strains. Nineteen of these isolates were defined as C. guilliermondii by DNA-based methods, either using a DNA probe specific for the C. guilliermondii clade (3) or through direct DNA sequencing of ribosomal DNA. The last isolate (B6832) is the type strain for C. carpophila (15).
TABLE 1.
Strains used in this study
| Strain and CDC no. or ATCC strain no. | Description (sample no.) or source | Genotypea |
|---|---|---|
| Brazilian outbreak strains (group 1) | ||
| 2004010326 | Patient isolate (1CG) | I, B |
| 2004010327 | Patient isolate (2CG) | I, B |
| 2004010328 | Patient isolate (3CG) | I, B |
| 2004010329 | Patient isolate (4CG) | I, B |
| 2004010330 | Patient isolate (5CG) | I, B |
| 2004010331 | Patient isolate (6CG) | I, B |
| 2004010332 | Patient isolate (7CG) | I, B |
| 2004010333 | Patient isolate (8CG) | I, B |
| 2004010334 | Patient isolate (9CG) | I, B |
| 2004010335 | Patient isolate (10CG) | I, B |
| 2004010336 | Patient isolate (11CG) | I, B |
| 2004010337 | Patient isolate (12CG) | I, B |
| 2004010338 | Patient isolate (13CG) | I, B |
| 2004010339 | Patient isolate (14CG) | I, B |
| 2004010340 | Patient isolate (15CG) | I, B |
| 2004010341 | Patient isolate (16CG) | I, B |
| 2004010342 | Patient isolate (17CG) | I, B |
| 2004010443 | Patient isolate (18CG) | I, B |
| 2004010444 | Patient isolate (19CG) | I, B |
| 2004010445 | Patient isolate (20CG) | I, B |
| 2004010446 | Isolate 21 from a nurse (21CG) | I, B |
| 2004010447 | Isolate 22 from a nurse (22CG) | I, B |
| 2004010448 | Isolate 23 from a nurse (23CG) | I, B |
| 2004010449 | Isolate 24 from a sphygmomanometer cuff (24CG) | I, B |
| 2004010450 | Isolate 25 from paper towel | I, B |
| South American control strains (group 2) | ||
| 20233.092 | São Paulo, Brazil | I, C |
| 20313.020 | Santa Fe, Argentina | I, C |
| 20319.018 | São Paulo, Brazil | I, C |
| 20336.020 | Bogota, Colombia | I, A |
| 20337.052 | Medellin, Colombia | I, C |
| 20337.091 | Medellin, Colombia | I, B |
| 20352.081 | Quito, Ecuador | III, F |
| 20361.066 | Buenos Aires, Argentina | III, D |
| 20416.031 | Bogota, Colombia | I, A |
| U.S. nonoutbreak control strains (group 3) | ||
| 2002015519 | I′, C | |
| 2002500017 | III, D | |
| 200201523/CAS00-0036 | II, I | |
| 200201524/CAS00-0037 | III, F | |
| 2002200337/B6222 | I′, C | |
| MAS92-0058 | I′, C | |
| MAS92-0116 | I′, ? | |
| MAS93-0313 | III, E | |
| MAS93-0386 | I′, B | |
| MAS93-0623 | II, G | |
| MAS93-0975 | I′, B | |
| MAS93-1057 | II, G | |
| LM17-95 | I′, C | |
| LM549-96 | I′, ? | |
| LM823-96 | II, H | |
| LM965-96 | I′, B | |
| B4346 | I′, B | |
| B4347 | I′, B | |
| KORG1 | I′, C | |
| B6832 | C. carpophila type strain | II, H |
| ATCC strains | ||
| ATCC 42715 | C. guilliermondii | I, A |
| ATCC 6260 | C. guilliermondii type strain | I, A |
Roman numerals refer to the RAPD group, and the letters refer to the HinFI type.
All 54 isolates were analyzed by randomly amplified polymorphic DNA (RAPD) fingerprinting using eight primers individually. Primers are given in Table 2. The four primers listed as UBC147, UBC194, UBC734, and UBC738 have been used previously in a C. guilliermondii outbreak investigation (12). These primers were originally described by Zeng et al. (22). The remaining four primers (Table 2) were from a panel of RAPD primers previously used for other species and found empirically to give fingerprint patterns (10). In addition, all isolates were analyzed by HinfI digestion of total genomic DNA. This technique for strain and Candida species identification was originally described by Fujita and Hashimoto (5).
TABLE 2.
Primers used in the RAPD analysis of 54 isolates
| Primer | Sequence |
|---|---|
| UBC147 (405624) | GTGCGTCCTC |
| UBC194 (405625) | AGGACGTGCC |
| UBC734 (405626) | GGAGAGGGAG |
| UBC738 (405627) | GGTGGGTGGT |
| RAPD2 (300960) | CACATGCCT |
| PAO3 | AGTCAGCCAC |
| M13core | GAGGGTGGCGTTCT |
| ERIC1R | ATGTAAGCTCCTGGGGATTCAC |
For RAPD profile analysis, DNA was prepared from fresh YEPD (1% yeast extract, 2% peptone, 1% dextrose) slant cultures using a Mo Bio Microbial DNA isolation kit (Mo Bio Laboratories, Inc., Solana Beach, CA) according to the recommendations of the manufacturer. For HinfI analysis, DNA was prepared from overnight 25-ml YEPD cultures using standard techniques as previously described (9). Basically, this consists of lysing spheroplasts, followed by proteinase K digestion, phenol extraction, and ethanol precipitation. Overnight digestions of genomic DNA with HinfI (Roche, Indianapolis, IN) at 37°C were electrophoresed on 1.0% agarose at 5 μg/lane, followed by ethidium bromide staining.
For RAPD profiling, PCR conditions were as previously described (9), using RAPD primers individually as listed in Table 2. Amplification conditions were as follows: 5-min denaturation at 95°C, 35 cycles (with 1 cycle consisting of 1 min at 95°C, 1 min at 25°C, and 1 min at 72°C), and a final 5-min extension at 72°C. The entire reaction mixture (20-μl total volume) was electrophoresed on individual lanes of a 2% Metaphor agarose gel (FMC Corp., Rockland, MA). Molecular size standards (number 9) were from Roche (Indianapolis, IN). RAPD fingerprint patterns were analyzed using Bionumerics (Applied Maths, Austin, TX) software.
Adoption of intervention measures.
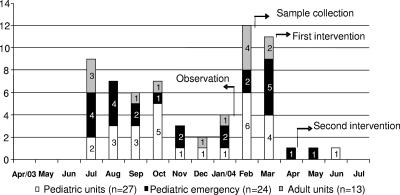
The first intervention measure, involving lectures for the medical and nursing teams of the pediatric units, was adopted in March 2004. The importance of complying with guidelines for infection control, including hand hygiene and techniques for blood collection as described in the hospital manuals, were the main topics discussed. Despite the adoption of these measures, some cases of C. guilliermondii-positive blood cultures were still observed in the pediatric units, which led the HICC to determine that blood samples should be drawn by a specialized professional. Members of the nursing staff, from different sectors and working different shifts, were later selected for the task of blood collection. In April 2004, the second intervention procedure, involving the theoretical and practical training of these workers, was established. Training included all the steps of the technique of drawing blood samples for culture, related to hand hygiene, aseptic preparation of the patient's skin, preparation of the material for collection, and additional care measures during the procedure.
RESULTS
C. guilliermondii was identified in 64 (43%) of the 149 blood cultures between June 2003 and July 2004. The samples were from patients hospitalized in different wards but were concentrated in pediatric units (Fig. 1). A total of 51 (79.7%) blood cultures positive for C. guilliermondii were obtained from pediatric units: 24 (47.1%) were from the Pediatric ED, and 27 (52.9%) from other pediatric units. The remaining 13 (20.3%) positive blood cultures were derived from adult patients. Results from the clinical and epidemiological investigation of the cases suggested a pseudo-outbreak, since most of the patients had not been submitted to invasive procedures or exposed to factors which could be associated with Candida infections. The patients were, furthermore, mainly from pediatric emergency wards, and many of them did not show signs or clinical symptoms of severe infection and were not hospitalized. No deaths occurred, even when the patients were not treated. During the first stage of the study, 15 sessions of blood collection involving different members of the medical and nursing staff were observed. Analysis of the forms filled in during each session showed the existence of flaws in the procedures of hand washing and asepsis by the health care workers, as well as during preparation of the patient's skin for drawing blood (Table 3). These results confirm the lack of adherence to guidelines for blood collection established by the hospital.
FIG. 1.
Distribution of C. guilliermondii cases isolated at the Hospital São Paulo, São Paulo, Brazil, from April 2003 to July 2004 and intervention measures. The numbers on the y axis and in the bars are the numbers of cases.
During the pseudo-outbreak, C. guilliermondii was isolated during culture of blood samples from different units of the hospital, with important concentration of patients from pediatric units (51 of 64; 79.7%) which included the Pediatric ED. The standard technique for the collection of blood samples is puncture of the cubital vein at the elbow. In the patients where this method was not possible, blood was obtained from a scalp vein.
The microbiological investigation of materials and surfaces showed the presence of C. guilliermondii in the sphygmomanometer cuff, paper towels, and sink drain at the Pediatric ED. The fungus was not detected in samples of cotton, gauze, tape, antiseptic solution, from the tray, from the surface of the soap tray or sink, and from the vials used for blood collection. Analysis of the samples collected from the 32 members of the nursing staff showed that 20 (79%) were colonized with fungi, and that in 7 cases (21%), the colonization was due to C. guilliermondii. As part of the intervention measures, the professionals were sent to be evaluated by a specialized physician, who concluded that none of the individuals displayed skin or nail fungal infections.
The pseudo-outbreak was completely resolved when blood samples began to be collected only by members of the nursing team assigned to this task and adequately trained according to institutional standards.
Subtyping of C. guilliermondii isolates. (i) Group 1 (Brazilian pseudo-outbreak strains).
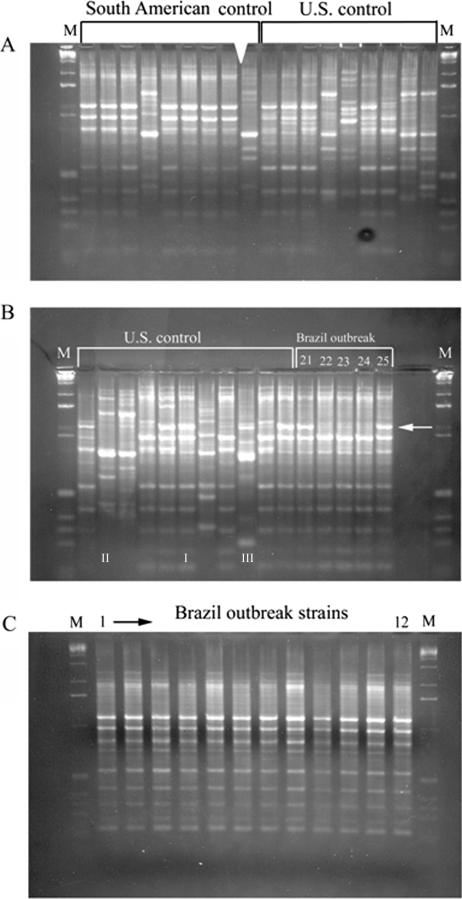
For seven of the eight RAPD primers, we observed no reproducible differences among the 25 isolates. Each RAPD primer gave a different fingerprint profile, which was shared by all isolates in this group (data not shown). However, all primers gave different patterns for the nonoutbreak isolates as discussed below. We conclude, therefore, that the primers can detect differences between strains and that group 1 outbreak strains are highly related. One RAPD primer (UBC738 or 405627) could distinguish between patient strains and some of the environmental strains (isolates 21 to 25). This is shown in Fig. 2. Specifically, isolates 22, 23, and 24, corresponding to two isolates from two nurses and a blood pressure cuff (Fig. 2B), lack a major band of approximately 650 base pairs that was seen in the other blood and environmental isolates. The other isolates from nurses and paper towels gave patterns matching the patients' isolates. These experiments were repeated using different DNA preparations and gave identical results.
FIG. 2.
RAPD fingerprints of Brazilian cluster strains using primer UBC738 or 405627. (A) South American control and U.S. control strains. (B) U.S. control and Brazilian outbreak strains 2004010446 to 2004010450 (21 to 25, respectively, in text). The white arrow indicates the absence of bands for isolates 22 to 24. (C) Brazilian outbreak strains 2004010326 to 2004010337 (strains 1 to 12, respectively, in text). I to III in the lanes show examples of the three representative groups. M, molecular size markers.
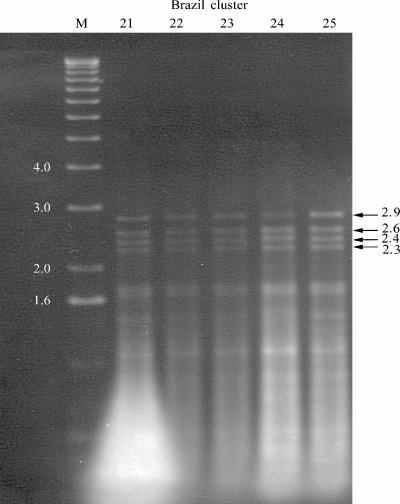
HinfI digestion resulted in four major bands at 2.9, 2.6, 2.4, and 2.3 kb; representative patterns are shown in Fig. 3, which shows strains 21 to 25 (environmental strains). All 25 strains gave identical patterns, which has been subsequently termed pattern B. This gives additional support for the concept that outbreak strains are highly related.
FIG. 3.
HinfI digestion pattern B for five environmental isolates in the Brazilian cluster strains. Molecular sizes (in kilobases) are shown at the sides of the gel.
(ii) Analysis of all groups.
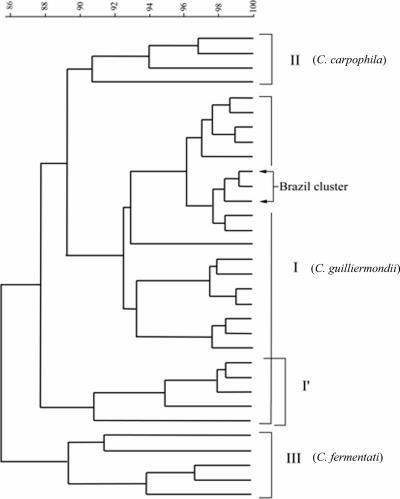
All eight RAPD primers consistently gave the same results. Although the patterns were different for each primer, there were no inconsistencies between profile types. The results for primer UBC738 are shown in Fig. 2, with South American control strains shown in Fig. 2A, U.S. control strains in Fig. 2A and B, and Brazilian strains in Fig. 2B and C. These data were used as part of a cluster analysis with primer UBC147 (Fig. 4) and can be summarized as follows. RAPD profiles cluster into three major groups, termed I, II, and III. We observed a subgroup of group I, termed I′. Members of the I′ group differ by one or two bands from group I. All Brazilian pseudo-outbreak strains were found to be members of group I (only three representative strains are shown in the cladogram). Group I members also include ATCC 42715 and strain ATCC 6260, the type strain for C. guilliermondii. Group II includes isolates similar to Candida (Torulopsis) carpophila (5, 15). Group III includes strains with HinfI patterns similar to those of Candida (Torula) fermentati (15) and was consistent with DNA sequence information obtained from selected members of this group. We conclude, therefore, that RAPD fingerprint profiles can distinguish between distinct taxonomic groups at the species level.
FIG. 4.
Cluster analysis of RAPD fingerprint patterns obtained with primers UBC147 and UBC738 by the unweighted-pair group method using average linkages. The 29 nonoutbreak strains and three examples of the outbreak strains are shown. Roman numerals I to III correspond to the three major taxonomic groups. Percent relatedness is shown on the horizontal axis.
The HinfI profiles were found to subdivide members of RAPD groups I to III further, consistent with the partitioning of the three groups. We observed a total of eight different HinfI patterns, termed A to H (data not shown). All RAPD group I strains had pattern A, B, or C. RAPD group II and III strains had patterns E to H. Each strain had a distinct, individual pattern. All 25 Brazilian outbreak strains were HinfI pattern B (Fig. 3). Therefore, their combined RAPD/HinfI genotype was I, B. The individual genotypes for all 54 strains in this study are given in Table 1. In addition, we observed that HinfI patterns A, B, and C were related by the simple gain or loss of a HinfI site. For example, pattern B (four bands) is related to pattern A (three bands) by the loss of the 2.9-kb fragment. This could be explained by the gain of a site in this fragment. These data are consistent with the concept of a single taxonomic group (group I) with single nuclear polymorphisms within the HinfI fragments. HinfI patterns D to H were associated with RAPD groups II and III. These patterns were more complex and presumably not just the simple gain or loss of a restriction site. Again, this is consistent with the findings that RAPD groups II and III are taxonomic entities distinct from RAPD group I and from each other. The segregation of these isolates into three HinfI patterns (A to C) can further subdivide conventionally indistinguishable C. guilliermondii strains. Approximately 50% (7 of 16) nonoutbreak RAPD group I control strains showed HinfI pattern B, with the remainder divided between patterns A and C.
DISCUSSION
Although C. guilliermondii is known to be invasive, hospital outbreaks are very rare and few reports are known. A pseudo-outbreak of C. guilliermondii infection, due to the contamination of heparin vials in a neonatal intensive care unit (17 babies) (21), and a hospital outbreak of C. guilliermondii resulting in invasive infection (n = 5) have been previously reported (12). Nonfungal pseudo-outbreaks of positive blood cultures have been found associated with contamination of the environment in which the blood was collected, use of contaminated gloves, disinfectants, collection tubes, or blood culture media and during sampling with an automated blood culture system (6, 11, 14).
Microorganisms on the skin of the patients or health care workers may be introduced into blood sample vials due to lack of adherence to good practices for control of hospital infection, with results that can be measured in both financial and human terms. One study showed that contaminated blood cultures can result in an increase of around 4.5 days in hospital stay and of more than $5,000 in the cost of treatment (13). According to standards published by the American Society for Microbiology, the rates of contamination of blood cultures should not exceed 3% (17), but it is not possible to eliminate all suspicious false-positive results. Some suspicious contaminations, in fact, may be associated with transitory bacteremias (17).
A number of factors have previously been shown to be important in interpreting the results of blood culture. These include training the workers who collect blood, selection and preparation of the puncture site, use of equipment for individual protection during blood collection, and volume of blood collected. A study by the College of American Pathologists showed that when blood is collected by nontrained personnel, a culture contamination rate of 77% is observed (14). The lowest contamination rates, on the other hand, are seen in facilities where specialized personnel are available (14).
Selection of the puncture site has a significant impact in the possibility of contamination of the blood culture. Collection of blood from areas involving peripheral or central vascular devices result in high contamination rates. Such invasive devices are maintained during long periods of time and are thus susceptible to colonization by microorganisms, which multiply and accumulate around the invasive ports and can thus be pulled into blood specimens collected from those sites and introduced into the blood culture vials (4).
Aseptic preparation of the puncture site is without question the most important factor in the collection of blood samples. Several antiseptics may be used for preparing the patient's skin, as long as compatibility of the products and the length of time they need to work are considered. Contamination of the puncture site is most likely when the access points are difficult to locate and the professional needs to repalpate the already aseptically prepared site (4).
The early identification of a pseudo-outbreak has great importance, as the patients may be unnecessarily treated, submitted to invasive procedures, have their hospitalization period extended, and have the real cause of their symptoms overlooked (11).
It was concluded from the subtyping results that the pseudo-outbreak strains from the patients are similar and are related to some of the environmental strains. This supports the hypothesis that this cluster originated from one or a limited number of common sources. This is consistent with the epidemiological findings.
It can be concluded that all the intervention measures adopted were important for resolving the pseudo-outbreak, but the single most important factor was the selection and training of blood collection personnel, showing the importance of the involvement of the health care workers, together with administrative measures.
Acknowledgments
We are grateful to Daniela Bicudo, Fernanda Crossera Parrera, Marcelo Abramczyk, Thais Guimaraes, and the nurses of the Division of Nursing of the Hospital São Paulo, UNIFESP, for their work in the control of the pseudo-outbreak.
Footnotes
Published ahead of print on 17 January 2007.
REFERENCES
- 1.Colombo, A. L., M. Nucci, B. J. Park, S. A. Nouér, B. Arthington-Skaggs, D. A. da Matta, D. Warnock, and J. Morgan for the Brazilian Network Candidemia Study. 2006. Epidemiology of candidemia in Brazil: a nationwide sentinel surveillance of candidemia in eleven medical centers. J. Clin. Microbiol. 44:2816-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colombo, A. L., M. Nucci, R. Salomão, M. L. M. Branchini, R. Richtmann, A. Derossi, and S. B. Wey. 1999. High rate of non-albicans candidemia in Brazilian tertiary care hospitals. Diagn. Microbiol. Infect. Dis. 34:281-286. [DOI] [PubMed] [Google Scholar]
- 3.Elie, C. M., T. J. Lott, E. Reiss, and C. J. Morrison. 1998. Rapid identification of Candida species with species-specific DNA probes. J. Clin. Microbiol. 36:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ernst, D. J. 2004. Controlling blood-culture contamination rates. MLO 36:14-18. [PubMed] [Google Scholar]
- 5.Fujita, S., and T. Hashimoto. 2000. DNA fingerprinting patterns of Candida species using HinfI endonuclease. Int. J. Syst. Evol. Microbiol. 50:1381-1389. [DOI] [PubMed] [Google Scholar]
- 6.Grinbaum, R. S., T. Guimaraes, E. Kusano, N. Hosino, H. Sader, and R. F. Cereda. 2003. A pseudo-outbreak of vancomycin-resistant Enterococcus faecium. Infect. Control Hosp. Epidemiol. 24:461-464. [DOI] [PubMed] [Google Scholar]
- 7.Krcmery, V., and A. J. Barnes. 2002. Non-albicans Candida spp. causing fungaemia: pathogenicity and antifungal resistance. J. Hosp. Infect. 50:243-260. [DOI] [PubMed] [Google Scholar]
- 8.Krcmery, V. 1999. Candidemia in cancer patients. Risk factors and outcome in 140 episodes from a single cancer institution. Acta Chemother. 5:133-145. [Google Scholar]
- 9.Lott, T. J., R. E. Fundyga, M. E. Brandt, L. H. Harrison, A. N. Sofair, R. A. Hajjeh, and D. W. Warnock. 2003. Stability of allelic frequencies and distributions of Candida albicans microsatellite loci from U.S. population-based surveillance isolates. J. Clin. Microbiol. 41:1316-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lott, T. J., R. J. Kuykendall, S. F. Welbel, A. Pramanik, and B. A. Lasker. 1993. Genomic heterogeneity in the yeast Candida parapsilosis. Curr. Genet. 23:463-467. [DOI] [PubMed] [Google Scholar]
- 11.Maki, D. G. 1980. Through a glass darkly. Nosocomial pseudoepidemics and pseudobacteremias. Arch. Intern. Med. 140:26-28. [PubMed] [Google Scholar]
- 12.Masala, L., R. Luzzati, L. Maccacaro, L. Antozzi, E. Concia, and R. Fontana. 2003. Nosocomial cluster of Candida guilliermondii fungemia in surgical patients. Eur. J. Clin. Microbiol. Infect. Dis. 22:686-688. [DOI] [PubMed] [Google Scholar]
- 13.Schifman, R. B. 1998. Phlebotomists at risk. Mayo Clin. Proc. 73:703-704. [DOI] [PubMed] [Google Scholar]
- 14.Schifman, R. B., C. L. Strand, F. A. Meier, and P. J. Howanitz. 1998. Blood culture contamination: a College of American Pathologists Q-Probes study involving 640 institutions and 497134 specimens from adult patients. Arch. Pathol. Lab. Med. 122:216-221. [PubMed] [Google Scholar]
- 15.Vaughan-Martini, A., C. P. Kurtzman, S. A. Meyer, and E. B. O'Neill. 2005. Two new species in the Pichia guilliermondii clade: Pichia carribica sp. nov., the ascosporic state of Candida fermentati, and Candida carpophila comb. nov. FEMS Yeast Res. 5:463-469. [DOI] [PubMed] [Google Scholar]
- 16.Viscoli, C., C. Girmenia, A. Marinus, L. Collette, P. Martino, B. Vandercam, C. Doyen, B. Lebeau, D. Spence, V. Krcmery, B. De Pauw, and F. Meunier. 1999. Candidemia in cancer patients: a prospective, multicenter surveillance study by the Invasive Fungal Infection Group (IFIG) of the European Organization for Research and Treatment of Cancer (EORTC). Clin. Infect. Dis. 28:1071-1079. [DOI] [PubMed] [Google Scholar]
- 17.Weinbaum, F. I., S. Lavie, M. Danek, D. Sixsmith, G. F. Heinrich, and S. S. Mills. 1997. Doing it right the first time: quality improvement and the contaminant blood culture. J. Clin. Microbiol. 35:563-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wenzel, R. P., and C. Gennings. 2005. Bloodstream infections due to Candida species in the intensive care unit: identifying especially high-risk patients to determine prevention strategies. Clin. Infect. Dis. 41(Suppl. 6):S389-S393. [DOI] [PubMed] [Google Scholar]
- 19.Wingard, J. R. 1995. Importance of Candida species other than C. albicans as pathogens in oncology patients. Clin. Infect. Dis. 20:115-125. [DOI] [PubMed] [Google Scholar]
- 20.Wisplinghoff, H., T. Bischoff, S. M. Tallent, H. Seifert, R. P. Wenzel, and M. B. Edmond. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 9:309-317. [DOI] [PubMed] [Google Scholar]
- 21.Yagupsky, P., R. Dagan, M. Chipman, A. Goldschimied-Reouven, E. Zmora, and M. Karplus. 1991. Pseudo-outbreak of Candida guilliermondii fungemia in a neonatal intensive care unit. Pediatr. Infect. Dis. J. 10:928-932. [DOI] [PubMed] [Google Scholar]
- 22.Zeng, S., L. C. Wu, and P. F. Lehmann. 1996. Random amplified polymorphic DNA analysis of culture collection strains of Candida species. J. Med. Vet. Mycol. 34:293-297. [DOI] [PubMed] [Google Scholar]