Abstract
The human bocavirus (HBoV) was recently isolated from respiratory tract samples. Within a study collective of children with severe lower respiratory tract disease, the patients testing positive for HBoV (12.8%) had a higher rate of underlying cardiopulmonary disease. Viral loads in respiratory tract specimens varied from 102 to 1010 genome equivalents/ml.
In addition to established pathogens for lower respiratory tract infections, such as respiratory syncytial virus (RSV), influenza viruses, parainfluenza viruses, and adenoviruses, additional agents have been identified in recent years. This includes the human metapneumovirus (hMPV), coronaviruses, and the human bocavirus (HBoV). HBoV is a member of the family Parvoviridae, genus Bocavirus, and has been isolated from respiratory tract samples by large-scale molecular virus screening (1). So far, the virus has not been propagated in cell culture; there is no animal model. The incidence of viral detection in patients with lower respiratory tract infections is 3.1% to 10.3% (1, 4, 5, 6). The rate of coinfection with other viruses ranges from 17% to 55%. HBoV infections have been recorded worldwide in all age groups and show predominance in winter and spring. The patients may develop pneumonia and bronchitis with fever, cough, and peribronchial infiltrates, detected on chest X-ray studies. Hospitalization of adults is rare.
Our study included 94 hospitalized patients of <36 months of age (mean, 6 months) with assumed severe lower respiratory tract disease as defined by a high rate of oxygen therapy (68%). An underlying cardiac and/or pulmonary disease was present in 18%. In most cases RSV infection was the most important differential diagnosis (53.2%) because of the seasonal epidemiology and the clinical symptomatology including marked airway obstruction in 62%. We chose this patient collective as young children represent reportedly the age group of HBoV genome-positive patients exhibiting the severest symptoms. Among 75 patients investigated by chest X-ray studies, 72% had peribronchial and 23% had pneumonic infiltrates. Respiratory tract specimens collected from these children between November 2005 and April 2006 (76% nasopharyngeal washes, 21% tracheal secretions, 3% bronchoalveolar lavage [BAL] samples) were studied for adenoviruses, influenza A virus, influenza B virus, parainfluenzaviruses types 1 to 3, and RSV using antigen-specific immunofluorescence assays (IMAGEN respiratory screen; DAKO); for the hMPV using a PCR assay (2); and for HBoV using the PCR protocol described by Allender et al. (1). Specimens known to be positive for parvovirus B19 genotype 1 (n = 5), herpes simplex virus, human cytomegalovirus, Epstein-Barr virus, JC virus, adenovirus, papillomavirus, RSV, parainfluenza virus, influenza A virus, or influenza B virus (n = 3 for each virus) were analyzed as specificity controls.
Nucleic acid was extracted from 220 μl of each specimen by using a QIAamp virus Biorobot 9604 kit (QIAGEN, Hilden, Germany) and eluted with 60 μl of PCR-grade water. An aliquot of 5 μl was added to 15 μl of reaction mixture containing 3 mM MgCl2, a 0.5 μM concentration of each primer (1888 F, 542 R, and HBoV NP1 gene) (1), 0.15 μM fluorescein hybridization probe (GGAAGAGACACTGGCAGACAAC-fluorescein; TIB Molbiol, Berlin, Germany), 0.15 μM LC-Red 640 probe (LC-Red 640-CATCACAGGAGCAGGAGCCG), and 2 μl of enzyme mix (LightCycler FastStart DNA Master Hybridization Probes; Roche Applied Science, Mannheim, Germany). The experimental PCR protocol was as follows: an initial 10 min at 95°C for FastStart Taq polymerase activation, followed by 45 cycles of 2 s of denaturation at 95°C, 10 s of annealing at 54°C, and 15 s of extension at 72°C. Quantification of HBoV DNA was performed with a serial dilution of a plasmid standard containing the primer-spanning region of the NP1 gene. The amplicon of 354 bp was generated by PCR using DNA from an HBoV-positive specimen as a template and subsequently cloned into the vector pcDNA3.1/V5-His-TOPO (Invitrogen, Carlsbad, CA), resulting in the plasmid pcDNA-HBoV.
Of the 94 specimens included in this study, 26 (27.6%) were negative in all assays (Table 1); 50 (53.2%) were positive for RSV; 7 (7.4%) were positive for adenoviruses, influenzaviruses, or parainfluenzaviruses; and 5 (5.3%) were positive for the human metapneumovirus. HBoV could be detected in 12 (12.8%) specimens (83.3% were nasopharyngeal washes, 8.3% tracheal secretions, 8.3% BAL samples) of patients aged 1 to 30 months (mean, 7.8 months), five of whom were simultaneously infected with RSV (41.7%). In 3 of the 12 HBoV-positive patients, hospital-acquired infection was assumed as they developed respiratory symptoms after hospital treatment for other diseases for at least 4 weeks.
TABLE 1.
Clinical and virological findings 94 young children with assumed viral lung infections
| Virus | nb | Age (mo)c | No. of children with:
|
Chest X-ray results (no. of children)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pulmonary obstruction | Oxygen therapy | Rhinopharyngitis | Tonsillitis | Enteritisd | Underlying cardiopulmonary diseasee | Sample | Peribronchial infiltrates | Pneumonic infitrates | Dys- or atelectasis | Pleural effusion | |||
| None | 26 | 6.1 ± 6.8 | 15 (58%) | 16 (62%) | 11 (42%) | 0 | 2** (8%) | 8 (31%) | 22 | 15 (68%) | 5 (23%) | 3 (14%) | 3 (14%) |
| hMPV | 4 | 4.6 ± 3.7 | 2 (50%) | 2 (50%) | 2 (50%) | 0 | 0 | 0 | 4 | 3 (75%) | 1 (25%) | 1 (25%) | 1 (25%) |
| HBoV | 7 | 9.6 ± 9.3 | 2 (29%) | 4 (57%) | 4 (57%) | 0 | 0 | 3 (43%) | 6 | 3 (50%) | 2 (33%) | 0 | 0 |
| RSV | 44 | 5.1 ± 3.6 | 34 (77%) | 33 (75%) | 39 (89%) | 0 | 8*** (18%) | 2 (4.5%) | 30 | 24 (80%) | 5 (17%) | 4 (13%) | 0 |
| Othera | 7 | 8.1 ± 5.8 | 3 (43%) | 4 (57%) | 7 (100%) | 1 (14%) | 0 | 2 (29%) | 7 | 5 (71%) | 2 (29%) | 1 (14%) | 0 |
| RSV + HBoV | 5 | 5.4 ± 2.3 | 5 (100%) | 4 (80%) | 4 (80%) | 0 | 1* (20%) | 2 (40%) | 5 | 3 (60%) | 2 (40%) | 2 (40%) | 0 |
| RSV + hMPV | 1 | 5 | 1 (100%) | 1 (100%) | 1 (100%) | 0 | 0 | 0 | 1 | 1 (100%) | 0 | 0 | 0 |
| Total | 94 | 62 (66%) | 64 (68%) | 68 (72%) | 1 (1%) | 11 (12%) | 17 (18%) | 75 | 54 (72%) | 17 (23%) | 11 (15%) | 4 (5%) | |
The other viruses include the following (number of cases): adenovirus (1), influenza B virus (1), parainfluenzavirus (3), and enterovirus (1).
n, no. of children.
Mean ± SD.
The number of cases of coinfection with rotavirus is indicated as follows: *, 1; **, 2; ***, 3.
Values in boldface indicate a significant increase of underlying cardiopulmonary disease in HBoV genome-positive patients (P < 0.05).
HBoV was second only to RSV as the most frequent pathogen detected among children with assumed severe lower respiratory disease in our study. In agreement with previous studies, coinfection with other pathogens was common (1, 5), which may reflect the high prevalence of viral infections in young children and does not deny the pathogenic potential of HBoV. The share of HBoV genome-positive tracheal secretions in all HBoV genome-positive specimens (8.3%) was low compared to the share of tracheal secretion specimens included in this study (21%). This may indicate that tracheal secretion is less suitable for detection of HBoV than nasopharyngeal washes and BAL. The prevalence of HBoV found in this study is the highest reported so far. This result may be explained by the preselection of severely diseased young children. In contrast, previously reported studies included either stationary patients and outpatients or stationary patients with less severe lung disease. HBoV-positive patients had a higher rate of underlying cardiopulmonary disease (5/12) than all remaining patients (12/82; P < 0.05), suggesting that these patients might be more susceptible to the pathogen (Table 1). However, no increased rates of rhinopharyngitis, tonsillitis, or enteritis were observed. In the patient with concomitant RSV and HBoV infection, enteritis could be attributed to a coinfection with rotavirus. In summary, no clear difference was seen with regard to the clinical presentation (obstructive lung disease or oxygen requirement) and the chest X-ray findings. Thus, no HBoV-specific clinical symptoms can be defined.
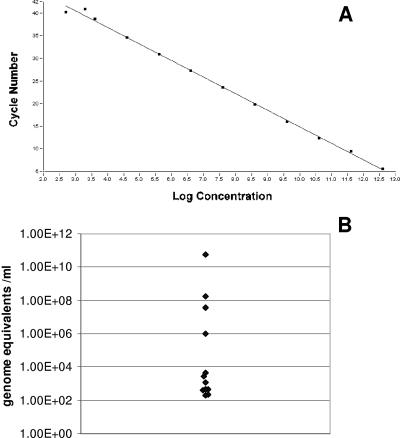
Cycle conditions for an HBoV-specific real-time PCR on the LightCycler instrument were established using primers and fluorophore probes as described above. The correlation between the crossing point and the log template concentration was determined with a dilution series of the standard quantification plasmid, pcDNA-HBoV. The analysis revealed a linear correlation (r = −1) between both parameters in a range between 103 and 1012 plasmid copies per ml of standard (Fig. 1A). To address the specificity of the assay, a panel of 35 samples positive for DNA viruses and respiratory viruses as described above was tested concomitantly. All samples were nonreactive by the HBoV-specific real-time PCR.
FIG. 1.
(A) Correlation between the crossing point and the log template concentration of an HBoV-specific real-time PCR using pcDNA-HBoV as a template. (B) HBoV viral load in respiratory specimens from 12 children with respiratory disease.
DNA preparations of the 12 specimens that tested positive for HBoV were retested with the new real-time PCR protocol, as were 25 HBoV-negative specimens. All specimens negative in the qualitative PCR were also negative in the real-time PCR. The 12 HBoV-positive specimens, in turn, were positive in the real-time PCR protocol, too. This indicates a high sensitivity and specificity of the novel real-time PCR assay. The quantification of these samples revealed viral loads ranging from 2 × 102 genome equivalents/ml to 5.6 × 1010 genome equivalents/ml (Fig. 1B) (mean, 5.6 × 109; median, 1.5 × 103). Low viral loads dominated. No correlation between viral load and the severity of clinical symptoms or patient's age was observed.
The data available so far indicate that HBoV may be responsible for a substantial share of respiratory tract infections in young children, particularly in those with underlying cardiopulmonary disease, narrowing the gap of infections with unknown etiology. Lu et al. (3) described an HBoV-specific real-time protocol using the exonuclease probe format. No absolute quantification of viral loads was given. The LightCycler PCR assay developed by us using the hybridization probe format allows the accurate detection and quantitative assessment of HBoV DNA in human specimens. Sensitivity and specificity of the assay are high. The specimens studied are dominated by low viral loads; only 25% of specimens had loads exceeding 104 genome equivalents/ml. Two hypotheses may explain the lack of correlation between the viral load and the clinical data. First, the nasopharyngeal washes, tracheal aspirates, and BAL samples were not collected by means of a standardized protocol for virus quantification but during routine procedures aiming at qualitative detection of viral particles, which may result in variable viral loads. Second, viral loads may change rapidly in the course of a disease, resulting in variable quantification results. The samples, however, had not been taken at predefined time points. Prospective studies based on quantitative HBoV detection are needed to specify the clinical impact of HBoV and to define the kinetics of the HBoV viral load.
Footnotes
Published ahead of print on 10 January 2007.
REFERENCES
- 1.Allender, T., M. T. Tammi, M. Eriksson, A. Bjerkner, A. Tiveljung-Lindell, and B. Andersson. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA 102:12891-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cote, S., Y. Abed, and G. Boivin. 2003. Comparative evaluation of real-time PCR assays for detection of the human metapneumovirus. J. Clin. Microbiol. 41:3631-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu, X., M. Chittaganpitch, S. J. Olsen, I. M. MacKay, T. P. Sloots, A. M. Fry, and D. D. Erdman. 2006. Real-time PCR assays for detection of bocavirus in human specimens. J. Clin. Microbiol. 44:3231-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma, X., R. Endo, N. Ishiguro, T. Ebihara, H. Ishiko, T Ariga, and H. Kikuta. 2006. Detection of human bocavirus in Japanese children with lower respiratory tract infections. J. Clin. Microbiol. 44:1132-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sloots, T. P., P. McErlean, D. J. Speicher, K. E. Arden, D. N. Nissen, and I. M. Mackay. 2006. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J. Clin. Virol. 35:99-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weissbrich, B. B., F. F. Neske, J. J. Schubert, F. F. Tollmann, K. K. Blath, K. K. Blessing, and H. W. Kreth. 2006. Frequent detection of bocavirus DNA in German children with respiratory tract infections. BMC Infect. Dis. 6:109. [DOI] [PMC free article] [PubMed] [Google Scholar]