Abstract
During February and March 2005, one of the largest national recorded outbreaks of severe acute gastroenteritis occurred in Nicaragua, affecting ≥64,000 individuals and causing ≥56 deaths, predominantly in children under 5 years of age. Through a nationwide laboratory-based study, stool samples were collected and investigated for rotavirus. Of 108 stool samples examined, 72 (67%) were positive for rotavirus. While 69% (50/72) of the positive samples were found in children less than 2 years of age, 50% (6/12) of the adult samples were positive. A mutated G4P[8] strain was the most commonly recognized strain (85%), followed by mixed G strains (8%) and G9P[8] (7%) strains. Phylogenetic analysis of the VP7 gene revealed that the G4 strains belonged to the emerging lineage Ic and was distantly related to the ST3 and VA70 G4 strains. Secondary structure predictions of the VP7 G4 protein revealed an insert of an asparagine residue in position 76, which, combined with additional mutations, surprisingly modified two downstream β-sheets at amino acid positions 80 to 85 and 115 to 119. The 2005 G4P[8] strain compared to a G4P[8] strain from 2002 had a substitution of an asparagine residue for threonine (Asn→Thr) at position 96 within antigenic region A, thus eliminating a potential glycosylation site. The mutated G4 virus was introduced in Nicaragua after 2002 and probably emerged from Brazil, Argentina, or Uruguay.
Rotavirus is the single most important etiologic agent of acute diarrhea mainly affecting children under the age of 5 (32). Each year human rotavirus (HRV) causes approximately 2 million hospitalizations and approximately 440,000 deaths, with the majority of the mortality in children from less industrialized countries (32).
It is estimated that all children will be infected at least once by the age of 5 years (5), which leads to protective immunity later in life. In adults rotavirus infections are usually asymptomatic, but there are reports of disease in elderly and immunocompromised patients (1, 4, 26, 28, 33).
Reports from different countries suggest an epidemiological shift of HRV strains over the years (20, 30). It seems that one specific variant or lineage within the serotypes might be responsible for outbreaks in well-defined geographic regions; for instance, the lineage Ic of the strain G4P[8] increased in prevalence in some cities of Argentina in 1998 and 1 year later in Paraguay (7, 9). The lineage Ic of the G4P[8] strain was also found to circulate in Italy in 1999 and 2000 (3), and during the rotavirus season in 1997 the emergence of a G4P[6] strain with a different RNA profile was observed in South Africa (34). Furthermore, in Nicaragua the uncommon strain G4P[6] was observed to circulate at very low frequency in 2001 and 2002 (12). The emergence of these novel strains and new lineages is due to different evolutionary mechanisms including gene rearrangements, accumulation of point mutations, and reassortment of genome segments (18).
During February and March 2005, one of the largest recorded outbreaks of severe acute gastroenteritis occurred in Nicaragua, affecting ≥64,000 individuals and causing at least 56 deaths, with children <5 years of age being most affected. We have carried out a molecular epidemiology study to investigate properties of the rotavirus strains associated with this unique and large epidemic outbreak. Our main observation was that the outbreak was associated with a mutated G4P[8] virus not previously detected in Nicaragua and most likely introduced from South America. The knowledge from this study may have implications for rotavirus prevention including vaccine introduction in Nicaragua.
MATERIALS AND METHODS
Geographic distribution of collected stool samples.
During February and March 2005, a total of 108 stool samples were collected through a laboratory-based survey of acute diarrhea cases, defined as three or more liquid stools over a 24-h period (12-14). The surveillance was performed in the main hospital of every city included in this study; therefore, most of the samples (55/108; 76.4%) came from moderate to severe cases. A portion of the samples (17/72; 23.6%) was collected in the laboratory of the Department of Microbiology of the National Autonomous University of Nicaragua-Leon serving mainly outpatients.
The stool specimens were stored at 4°C until transported to the Department of Microbiology of the National Autonomous University of Nicaragua-Leon. A suspension of 10% (vol/vol) phosphate-buffered saline (pH 7.0) was prepared for HRV antigen detection and two aliquots of stool-phosphate-buffered saline suspensions were stored at −20°C for further analysis.
The laboratories of the health care facilities from the health system participated in the survey. Samples were geographically representative from all over the country and collected as follows: Pacific region, 68% (including Leon, 40% [43/108]; Managua, 21% [23/108]; Chinandega, Granada, Masaya, and Jinotepe, 7% [8/108]); northern and central regions, 29% (including Matagalpa-Jinotega, 11% [12/108]; Esteli, 10% [11/108]; Madriz and Nueva Segovia, 5% [5/108]; and Chontales, 3% [3/108]); and the Caribbean coast, 3% (4/108). While age was not reported in 6% (7/108) of the cases, 40% (43/108) of the samples were identified as coming from children <12 months old, 82% (89/108) from children <5 years old, and 11% (12/108) from young people and adults (Table 1).
TABLE 1.
Distribution of G and P types of HRV strains isolated during an epidemic outbreak of acute gastroenteritis in Nicaragua 2005
| Age | No. of positive cases/no. of cases (%) | VP7 typing (no. of cases)a
|
VP4 typing (no. of cases)b
|
||
|---|---|---|---|---|---|
| G4 | G9 | Mix | P[8] | ||
| 0-5 mo | 11/11 (100) | 8 | 1 | 2 | 11 |
| 6-11 mo | 19/32 (59) | 14 | 4 | 1 | 19 |
| 12-23 mo | 20/29 (69) | 18 | 0 | 2 | 20 |
| 24-60 mo | 10/17 (59) | 9 | 0 | 1 | 10 |
| ≥12 yr | 6/12 (50) | 6 | 0 | 0 | 6 |
| Not determined | 6/7 (86) | 6 | 0 | 0 | 6 |
| Total | 72/108 (67) | 61 | 5 | 6 | 72 |
G types investigated: G1, G2, G3, G4, G8, G9, G10.
P types investigated: P[4], P[6], P[8], P[9], P[10], P[11].
Rotavirus antigen detection.
A commercial enzyme immunoassay (enzyme-linked immunosorbent assay kit K6020, IDEA Rotavirus; Dako Cytomation Ltd., United Kingdom) was used for detection of group A HRV in fecal samples following the manufacturer's instructions. The results were read visually and confirmed by absorbance readings.
RNA extraction.
Viral double-stranded RNA (dsRNA) was extracted from stool suspensions following the manufacturer's instructions using a QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany). A total of 60 μl of viral dsRNA was collected and stored at −20°C until RNA gel electrophoresis or reverse transcription was carried out.
Polyacrylamide gel electrophoresis.
RNA gel electrophoresis and negative silver staining were performed as previously described (35).
G and P multiplex genotyping.
G and P genotyping was performed as described previously (21). This method is a modification of the original methods described by Gouvea et al. for G typing and Gentsch et al. for P typing (15, 16) and includes random primers [pd(N)6] to produce the cDNA and modified G and P primers.
RT.
Reverse transcription (RT) was essentially carried out as described previously (19). Briefly, 28 μl of dsRNA was mixed with 50 pmol of random hexadeoxynucleotides [pd(N)6], denatured at 97°C for 5 min, and quickly chilled on ice for 2 min, followed by addition of one RT-PCR bead (Amersham Biosciences, United Kingdom) and RNase-free water to a final volume of 50 μl. The RT reaction was carried out for 30 min at 42°C to produce the cDNA used for genotyping and sequence analysis.
PCR.
A PCR mix was prepared using 5 μl of 10× Native plus PFU buffer (Stratagene, La Jolla, CA), 1 μl of 10 mM deoxynucleoside triphosphate (dNTP) mix (Applied Biosystems, Warrington, United Kingdom), 4 pmol of each consensus primer (VP7-F and VP7-R) (17), 2.5 U of Native DNA polymerase (Stratagene, La Jolla, CA), and RNase-free water to a final volume of 50 μl. The PCR was performed at 94°C for 2 min, followed by 35 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min, with a final extension of 72°C for 7 min.
The amplicons were analyzed by gel electrophoresis using 2% agarose gel and ethidium bromide staining, and a consensus region of 881 bp of the VP7 encoding gene was obtained. The PCR products were purified with spin column purification (QIAprep Spin Miniprep Kit; QIAGEN, Hilden, Germany) and the amount of DNA was determined by a NanoDrop ND-1000 UV-visible light spectrophotometer (Saveen Werner AB, Malmö, Sweden).
Cloning.
Purified PCR products were cloned into the pPCR-Script Amp SK(+) cloning vector, followed by transformation into XL10-Gold Kan ultracompetent cells according to the manufacturer's instructions (Stratagene, La Jolla, CA). The transformed bacteria were examined for recombinant plasmids by blue and white screening and PCR. The PCR mixture contained 2.5 μl of 10× PCR buffer (Invitrogen, Carlsbad, CA), 1 μl of 50 mM MgCl2 (Invitrogen, Carlsbad, CA), 5 pmol of each standard sequencing M13 forward and reverse primer, 2 μl of 2.5 mM dNTP (Invitrogen, Carlsbad, CA), 1 U of Taq DNA polymerase (Invitrogen, Carlsbad, CA), and RNase-free water to a final volume of 25 μl. The PCR was performed at 94° for 5 min, followed by 35 cycles of 94° for 30 s, 50°C for 30 s, and 72°C for 1 min, with a final extension of 72°C for 10 min.
Nucleotide sequencing.
One independent colony from each cloned sample containing the desired insert was collected for sequencing. However, before sequencing, the PCR products were cleaned from excess dNTP and primers by using ExoSap-IT (GE Healthcare, Chalfont St. Giles, United Kingdom), in a reaction mixture containing 2.5 μl of PCR product and 1 μl of ExoSap-IT enzyme. This reaction was performed in 37°C for 15 min, followed by inactivation of the enzyme at 80°C for 15 min. Nucleotide sequences were obtained according to the manufacturer's instructions using a DYEnamic Dye Terminator Kit (GE Healthcare, Chalfont St. Giles, United Kingdom), M13 forward and reverse primers, and a Mega BACE 500 automated sequencer (GE Healthcare, Chalfont St. Giles, United Kingdom). For accuracy, clones with inserts were sequenced four times in both the forward and reverse directions. Complete sequences were obtained by assembling overlapping contigs with DNASTAR (Madison, WI).
Sequence analysis.
Multiple sequence alignment of VP7 G4 proteins and nucleotide sequencing were performed, using the ClustalW algorithm, version 1.8, with default parameters on the European Bioinformatics Institute server. A phylogenetic analysis of the VP7 nucleotide alignment of the G4 and G9 strains was performed using the MEGA 3.1 software package. A tree was constructed using the neighbor-joining and Kimura two-parameter methods. The statistical significance of the relationships obtained was estimated by bootstrap resampling analysis (1,000 replications).
Secondary structure prediction.
Secondary structure prediction of the VP7 G4 protein was performed using the PSIPRED protein structure prediction server (University College London) (8, 29).
RESULTS
Epidemiological features of the outbreak.
In the middle of February 2005, an increase in reported cases of diarrhea was reported in the weekly epidemiological bulletin of the health system of Nicaragua (27). By the end of March, the morbidity rate was 117/100,000 inhabitants, and the mortality rate was 1/100,000 inhabitants (compared to a mortality rate in 2004 of 0.4/100,000). The morbidity and mortality rates were estimated to be 26% and 115% higher, respectively, than those reported for the same period in 2004. Most of the fatal cases (98%) occurred in children less than 2 years of age. The increase in morbidity and mortality for diarrhea in children was also observed in the beginning of February 2005 in two northern neighboring countries, El Salvador and Guatemala (2, 27).
High prevalence of rotavirus in children and adults.
Rotavirus antigen screening revealed that 72/108 (67%) samples examined from children and adults suffering from diarrhea were positive. Furthermore 69% (50/72) of the rotavirus-positive samples were obtained from children less than 2 years of age, and 14% (10/72) were from children between 2 and 5 years of age. Eight percent of the positive samples (6/72) were derived from young people and adults (12 to 72 years of age; average, 29 years of age), but more important, 50% (6/12) of all the young and adult patients were rotavirus positive (Table 1). Four of the six were from the same city, and three of those from the same neighborhood.
Nationwide rotavirus outbreak predominantly associated with G4P[8].
To obtain information about the G and P type distribution, all 72 HRV-positive isolates were processed for G and P genotyping. G4 strains were found in 85% (61/72) of the samples, G9 in 7% (5/72), and mixed G infections in 8% (6/72) of the samples, including G4-G1 (two samples), G4-G9 (two samples), G3-G9 (one sample) and G4-G3-G9 (one sample). To eliminate any possibility of cross-priming occurring in these mixed infections, all the mixed infections were reanalyzed by single-locus PCR with genotype-specific primers to confirm the presence of the constituent G types. Only one sample was not consistent with the multiplex RT-PCR and the single-locus PCR analysis. That sample was the G4-G8 mixed infection in the first analysis, but no result was observed in the single-locus PCR analysis with the G8 primer. However, a clear G4 genotype was observed with the G4 primer (data not shown). Thus, five mixed infections occurred and all in children less than 5 years of age.
The P typing revealed that all strains belonged to the P[8] genotype (data not shown). The most common G and P combinations were G4P[8], representing 85% (61/72), and G9P[8], with 7% (5/72). All mixed infections were P[8], and a G9P[8] isolate came from a child less than 1 year of age living in Esteli. The G4P[8] strains were detected in all age groups and all over the country, including the Pacific, central, and Caribbean regions (Table 2).
TABLE 2.
Description of isolates used for VP7 gene nucleotide sequencing
| Isolate | Age of patient | Isolation location | Genotype |
|---|---|---|---|
| Nic03/05 | 7 mo | Esteli | G9P[8] |
| Nic84/05 | 13 mo | Jinotega | G4P[8] |
| Nic118/05 | 22 yr | Carazo | G4P[8] |
| Nic181/05 | 12 mo | Chinandega | G4P[8] |
| Nic199/05 | 3 mo | Granada | G4P[8] |
| Nic19PG/05 | 1 mo | León | G4P[8] |
| Nic40PG/05 | 21 yr | León | G4P[8] |
| NicBH63/02 | 29 mo | León | G4P[6] |
To investigate if the G4P[8] isolates emerged from different ancestors and thus exhibited different RNA profiles or resulted from accumulations of minor point mutations from a dominant strain, G4 and G9 samples were analyzed by RNA polyacrylamide gel electrophoresis as described previously (35). All analyzed samples had long RNA profiles, and furthermore, there were no apparent differences in RNA profiles from samples collected in different geographic settings or belonging to different G types (G4P[8] versus G9P[8]) (data not shown).
Nucleotide sequence analysis.
Seven G4 strains, including a sample isolated in 2002, representing different age groups and geographic locations, were selected for VP7 sequence analysis. In addition a G9P[8] strain from Esteli was sequenced (Table 2).
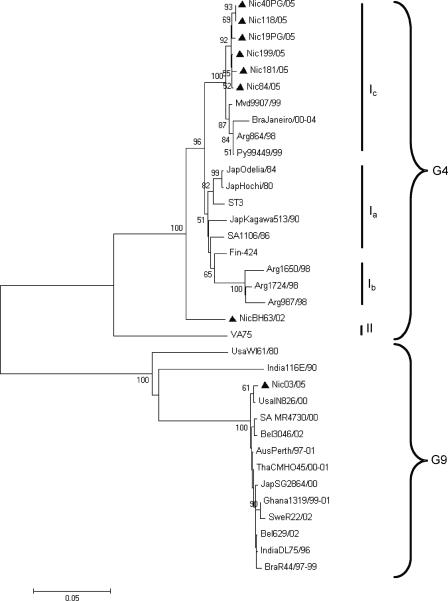
Bioinformatic analysis revealed that the G9P[8] strain (Nic03/05 [the year of isolation is indicated after the slash]) circulating in Esteli during the 2005 outbreak shared >99% homology at the nucleotide level with strains belonging to common G9 strains and particularly with strain IN826 (USAIN826/00) circulating in the United States in the years 2000 to 2001 (25) (Fig. 1). The G4P[6] virus (NicBH63/02) that circulated with very low (6%) frequency in 2002 in Nicaragua (12, 13) shared 94% homology at the nucleotide level with the G4P[8] strains circulating during the outbreak. Nucleotide sequence alignment of the VP7 gene from the G4P[8] strains (Table 2 and Fig. 2) indicated that they all belonged to the same cluster and shared >99% homology at the nucleotide level with strains isolated in Argentina, Uruguay, and Brazil between 1996 and 2004.
FIG. 1.
Phylogenetic analysis of VP7 nucleotide sequences of G4 and G9 strains. The tree was constructed using the Kimura two-parameter and neighbor-joining methods using MEGA 3.1 software. Bootstrap values are shown at the branch nodes (values of <50% are not shown). The Nicaraguan strains are marked with a bold triangle and the lineages (I) and genotypes (G) are indicated at the right. The number of substitutions per site is indicated by the scale bar. Abbreviations for locations: Aus, Australia; Bel, Belgium; Bra, Brazil; Fin, Finland; Mvd, Montevideo (Uruguay); Nic, Nicaragua; Jap, Japan; Py, Paraguay; SA, South Africa; Swe, Sweden; Tha, Thailand.
FIG. 2.
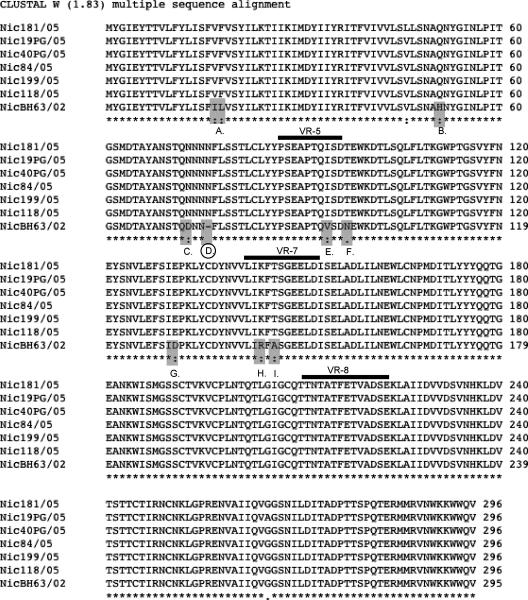
Unique asparagine insert in G4P[8] virus from 2005 in Nicaragua. Amino acid alignment of the rotavirus VP7 gene using ClustalW, version 1.8, with default parameters. The alignment reveals an asparagine insert at position 76 in the 2005 Nicaraguan isolates, which also is present in isolates from Argentina (1996 to 1998), Uruguay (1999), and Brazil (2000 to 2004). *, conserved residue; :, position with conserved substitutions; ., position with semiconserved substitutions.
Mutations in the G4 VP7 gene modify downstream β-sheets.
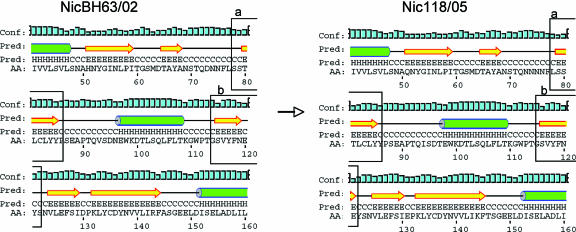
Alignment of G4 VP7 amino acids revealed a unique insertion of an asparagine residue at position 76 in the Nicaraguan strains compared to reference strains (Fig. 2). This insert is adjacent to the glycosylation motif Asn-X-Thr at residues 69 to 71, a site conserved in most human G4 strains (22). In addition to the asparagine insert, several other mutations were identified (Table 3). To investigate if these mutations affected the structure of the VP7 protein, a secondary structure prediction was performed using the PSIPRED protein structure prediction server (University College London). The secondary structure predictions revealed that the insertion of an asparagine together with the additional mutations (Table 3) resulted in two minor structural modifications, by altering two downstream β-sheets at amino acid positions 80 to 85 and 115 to 119 (Fig. 3).
TABLE 3.
Amino acid evolution in rotavirus G4 VP7 in Nicaragua between 2002 and 2005
| Location in Fig. 4 alignment | Position (aa)a | Amino acid (characteristics) in G4 strain from Nicaragua
|
|
|---|---|---|---|
| NicBH63/02 (2002) | Nic118/05 (2005) | ||
| A | 16 | I (neutral, nonpolar) | V (neutral, nonpolar) |
| 17 | L (neutral, nonpolar) | F (neutral, nonpolar) | |
| B | 51 | H (basic, polar) | Q (neutral, polar) |
| C | 73 | D (acidic, polar) | N (neutral, polar) |
| D | 76 | N (neutral, polar) | |
| E | 94 | V (neutral, nonpolar) | I (neutral, nonpolar) |
| F | 97 | N (neutral, polar) | T (neutral, polar) |
| G | 131 | D (acidic, polar) | E (acidic, polar) |
| H | 144 | R (basic, polar) | K (basic, polar) |
| I | 146 | A (neutral, nonpolar) | T (neutral, polar) |
The residue number is shifted 1 aa in the 2005 G4 VP7 sequence from position 76 in comparison to the 2002 strain.
FIG. 3.
Amino acid insertions and substitutions affect downstream β-sheets in VP7. Schematic representations of secondary structure predictions, using the PSIPRED protein structure prediction server (8). Both panels depict aa 41 to 160. The residue number is shifted 1 aa in the G4 VP7 sequence (Nic118/05) from position 76 compared to the 2002 strain (NicBH63/02). (Boxes a) Beginning at amino acid 80, the 6-amino-acid β-sheet in the 2002 isolate has mutated into a 7-amino-acid β-sheet in the 2005 isolate. (Boxes b) Located in the vicinity of amino acid 120, the 5-amino-acid β-sheet in the 2002 isolate has evolved into a 6-amino-acid β-sheet in the 2005 isolate. Conf, confirmed; Pred, predicted.
The G4P[8] virus of the 2005 outbreak shared amino acid similarities in antigenic VP7 regions with strains found in South America.
The amino acid substitutions in the G4P[8] strains influenced three antigenic sites described for VP7 (10, 11): site A, amino acids (aa) 87 to 96 (variable region 5 [VR-5]); site B, aa 142 to 152 (VR-7); and site C (VR-8), aa 211 to 223 (Table 4 and Fig. 4). The 2002 (NicBH63/02) and 2005 (Nic118/05) Nicaraguan G4P[8] strains were found to be identical in antigenic region C, while two amino acid substitutions had occurred in regions A (Val→Ile and Asn→Thr) and B (Arg→Lys and Ala→Thr) (Tables 3 and 4). Most interesting was the observation that the amino acid sequences in all three antigenic regions of G4P[8] strains isolated in Argentina, Uruguay, and Brazil between 1998 and 2004 were identical to the G4P[8] virus isolated in Nicaragua in 2005 but not in 2002.
TABLE 4.
The VP7 amino acid sequences in the antigenic regions of the G4P[8] outbreak virus are identical to those of the G4 viruses isolated in Argentina (1998), Uruguay (1999), and Brazil (2000 to 2004)
| Protein identifier | Strain | Antigenic region in VP7a
|
||
|---|---|---|---|---|
| Site A (aa 87 to 96, VR-5) | Site B (aa 142 to 152, VR-7) | Site C (aa 211 to 223, VR-8) | ||
| Nic 118/05 | S E A P T Q I S D T | I K F T S G E E L D I | N T A T F E T V A D S E K | |
| NicBH63/02 | - - - - - - V - - N | - R - A - - - - - - - | - - - - - - - - - - - - - | |
| AAL99429 | Arg864/98 | - - - - - - - - - - | - - - - - - - - - - - | - - - - - - - - - - - - - |
| AAN03765 | Mvd9907/99 | - - - - - - - - - - | - - - - - - - - - - - | - - - - - - - - - - - - - |
| AAX54691 | BraJaneiro/00-04 | - - - - - - - - - - | - - - - - - - - - - - | - - - - - - - - - - - - - |
| ST3 | - - - - - - - - - - | - R - V F - - - - - - | - - - - - - - - - - - - - | |
| AAA47395 | VA75 | - - - - - - - - - N | - - - A - - - - - - - | - V - - - - M - - - - - - |
FIG. 4.
Amino acid alignment using ClustalW, version 1.8, with default parameters on the European Bioinformatics Institute server of the rotavirus VP7 gene showing the number of amino acid substitutions in the 2005 Nicaraguan isolates compared to the isolate from 2002. The substitutions occurring at positions A to I are all conserved or semiconserved. The three important antigenic sites (VR-5, aa 86 to 101; VR-7, aa 142 to 152; and VR-8, aa 211 to 223) are underlined. The location of the asparagine insert at position 76 is indicated with a circle. *, conserved residue; :, position with conserved substitutions; ., position with semiconserved substitutions.
Furthermore, when the reference strains ST3 (G4P[6] subtype A), isolated in the 1970s, and VA70 (G4P[8] subtype B), isolated in 1975, were compared, the 2005 G4P[8] strain (Nic118/05) was found to be identical to the ST3 strain in antigenic regions A and C while having substitutions in all the regions compared to the VA70 stain (Table 4).
The G4P[8] strain of 2002 (NicBH63/02) was identical to strain ST3 in antigenic region C, while having substitutions in regions A and B. Compared to the VA75 strain, the G4P[8] 2002 strain as well as the G4P[8] strain isolated in 2005 had substitutions in all the compared regions. However, the VA75 strain and the G4P[8] strain isolated in 2002 have two identical substitutions compared to the 2005 G4P[8] strain, where there is one substitution of an asparagine residue for a threonine (Asn→Thr) at position 96 within antigenic region A, thus eliminating a potential glycosylation site (Table 4).
DISCUSSION
In this study we report on molecular characterization of the emerging and mutated G4P[8] strain and the first observation of G9 rotavirus in Nicaragua, both identified during one of the largest recorded outbreaks of severe rotavirus diarrhea in the history of Nicaragua. Our investigation showed that the increased frequency of diarrheal episodes during the outbreak was due mainly to G4P[8] virus, which accounted for 85% of the rotavirus-positive cases. Unfortunately, no virological data were available regarding the mortality of the patients investigated in this study. However, it is most reasonable to believe that a portion of the fatal cases seen in the outbreak were associated with rotavirus G4P[8], since this strain was the most common genotype identified in cities where high mortality was reported (Madriz, Bluefields [autonomous region of the Atlantic coast], Nueva Segovia Granada, Jinotega, and Leon) (27). While the G4P[8] strain was dominating in this large outbreak, it has previously not been observed in Nicaragua. It was not found in studies carried out in 1994 or in 2001, 2002, or 2003, but the uncommon variant G4P[6] was circulating at low (5%) frequency in 2001 and 2002 (13).
Sequence analysis of the VP7 gene from G4P[8] strains circulating in Argentina in 1998 and 1 year later in Paraguay and Uruguay demonstrated that an emerging lineage of this serotype was circulating in those countries (6, 7, 9). Interestingly, the G4P[8] virus identified in this outbreak shares >99% homology at the nucleotide level with strains isolated in Argentina, Uruguay, and Brazil from 1998 to 2004 and belongs to the G4 lineage Ic (6, 7, 36). This may indicate that introduction of the mutated and highly virulent G4P[8] variant in Nicaragua probably originated from the South American region. In support of this suggestion is the finding that the G4P[8] outbreak strain is less related to strains Hochi and Odelia isolated in Japan in 1980 and 1984, respectively, and to the South African G4 strains isolated in 1986 and the Nicaraguan G4 strains isolated in 2002 (13, 24, 31). The outbreak of G4P[8] virus in 2005 may thus have been a result of infection of a population without preexisting immunity. The fact that 50% of the examined adults were symptomatically infected with G4P[8] may indicate that the G4P[8] virus has been absent for some time in Nicaragua or that the virus more recently has mutated. The Nicaraguan G4 VP7 strains all had a unique asparagine residue insertion at position 76, an insertion also found in strains isolated in Argentina, Uruguay, and Brazil during 1998 to 2004. This finding also supports the suggestion of an emergence of G4P[8] virus from South America to a highly susceptible population in Nicaragua. Another, albeit less likely, explanation is that the G4P[8] outbreak strain may have originated from reassortment events between G4P[6] and G1P[8] both circulating at low (5%) and high (45%) frequencies, respectively, during 2001 and 2002 (13).
The asparagine insertion at position 76 in the VP7 gene is close to the glycosylation motif Asn-X-Thr at residues 69 to 71, a site conserved in most human G4 strains, and it cannot be ruled out that the insertion influences glycosylation and, hence, alters antigenic properties. Residue 76 is located in a hydrophilic region, and an insertion of an asparagine in this area may cause an increase in hydrophilicity in the region (6). Most surprising was the observation that the asparagine insertion together with other mutations resulted in two minor structural modifications by altering two downstream β-sheets in amino acid positions 80 to 85 and 115 to 119 (Fig. 3). The observation that the mutations affected downstream β-sheets is interesting and warrants further investigation.
The amino acid substitutions that occurred between 2002 and 2005 in the Nicaraguan strains influenced the amino acid composition of important antigenic sites (10, 11). As illustrated in Table 3, the substitutions are located at position 94 (Val→Ile) in antigenic region A and positions 144 (Arg→Lys) and 146 (Ala→Thr) in antigenic region B. Furthermore, we found that G4P[8] strains isolated in Brazil, Uruguay, and Argentina between 1998 and 2002 had amino acid sequences in antigenic regions A to C that are identical to those of the G4P[8] strain isolated in this outbreak but different from the Nicaraguan strain isolated in 2002. These findings suggest that the reemergence of G4 strains in Nicaragua could be due to immune evasion as a consequence of altered antigenicity conferred not only by the asparagine residue insertion in VP7 at position 76 but also by the substitution of the asparagine (Asn→Thr) residue at position 96 in antigenic region A, which eliminated an N-linked glycosylation site and thus possibly altered antigenicity (11). An older population, presumably with preexisting immunity but with a high incidence of HRV infection due to immune evasion, has also been suggested by Iturriza-Gomara et al. in a report describing a G2 strain causing gastroenteritis between 1995 and 1998 in the United Kingdom (20, 35).
It has previously been observed that the antigenic regions in the VP7 protein, even though distant in the linear molecule, interact closely together in the folded form of the VP7 molecule and thus influence, for instance, antibody binding and immune responses (11). Indeed, a single amino acid substitution in VP7 and predominantly at antigenic regions A, B, and C has been observed to alter antigenicity (22). Thus, it cannot be ruled out that the identified mutations result in a mutated virus displaying a different molecular makeup, conferring an increased virulence enabling this particular virus strain to escape neutralization by G4 antisera. Indeed, in support for this hypothesis, it has been suggested by Iturriza-Gómara et al. that the epidemic reemergence of G2 rotavirus strains in Taiwan was due to immune evasion as a consequence of altered antigenicity conferred by an amino acid substitution at position 96 in antigenic region A of the VP7 gene (17, 37). Interestingly, an identical substitution seen in the Nicaraguan strain (Asn→Thr) at position 96 was also observed in G4P[6] symptomatic strains isolated from neonates in South Africa (31). This suggests, but does not prove, that substitution of an asparagine to a threonine at position 96 may lead to increased virulence as seen in G4P[8] strains of this outbreak. Another observation in favor of increased virulence through immune evasion is the fact that 50% of the adults with diarrhea were rotavirus positive. However, it cannot be ruled out that the lack of immunity might have been associated with the P type.
Apart from El Salvador, the G9P[8] virus had not previously been detected in Central America (2, 12). In this study it represented 7% of the HRV-positive cases in children attending the health care centers of Managua and Esteli. An interesting observation was that no apparent difference in RNA profiles from samples collected in different geographic settings or belonging to different G types (G4P[8] or G9P[8]) was found. There is a possibility that the G9P[8] strain examined might have originated from the United States because of its high nucleotide homology (>99%) with strain IN826 (USAIN826/00) isolated in Indianapolis (Indiana) in 2000 to 2001. In support of this explanation is also the fact that both strains share a long RNA profile (25). Considering that serotype G9 is emerging and may present a progressive increase in occurrence, as happened in Australia from 1997 until 2001 (23), it is reasonable to suggest that this G type strain will continue to increase in Nicaragua and elsewhere in the Central American region.
Acknowledgments
This study was supported by grants from the Swedish International Development Cooperation Agency (grant 0.75007109) and by the Swedish Research Council (grant 10392).
Footnotes
Published ahead of print on 17 January 2007.
REFERENCES
- 1.Anderson, E. J., and S. G. Weber. 2004. Rotavirus infection in adults. Lancet Infect. Dis. 4:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrus, J. (ed). June 2005. Immunization newsletter, vol. 27. Pan American Health Organization, Washington, DC.
- 3.Arista, S., G. M. Giammanco, S. De Grazia, C. Colomba, and V. Martella. 2005. Genetic variability among serotype G4 Italian human rotaviruses. J. Clin. Microbiol. 43:1420-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Awachat, P. S., and S. D. Kelkar. 2006. Dual infection due to simian G3-human reassortant and human G9 strains of rotavirus in a child and subsequent spread of serotype G9, leading to diarrhea among grandparents. J. Med. Virol. 78:134-138. [DOI] [PubMed] [Google Scholar]
- 5.Bern, C., J. Martines, I. de Zoysa, and R. I. Glass. 1992. The magnitude of the global problem of diarrhoeal disease: a ten-year update. Bull. W. H. O. 70:705-714. [PMC free article] [PubMed] [Google Scholar]
- 6.Berois, M., S. Libersou, J. Russi, J. Arbiza, and J. Cohen. 2003. Genetic variation in the VP7 gene of human rotavirus isolated in Montevideo-Uruguay from 1996-1999. J. Med. Virol. 71:456-462. [DOI] [PubMed] [Google Scholar]
- 7.Bok, K., D. O. Matson, and J. A. Gomez. 2002. Genetic variation of capsid protein VP7 in genotype g4 human rotavirus strains: simultaneous emergence and spread of different lineages in Argentina. J. Clin. Microbiol. 40:2016-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryson, K., L. J. McGuffin, R. L. Marsden, J. J. Ward, J. S. Sodhi, and D. T. Jones. 2005. Protein structure prediction servers at University College London. Nucleic Acids Res. 33:W36—W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coluchi, N., V. Munford, J. Manzur, C. Vazquez, M. Escobar, E. Weber, P. Marmol, and M. L. Racz. 2002. Detection, subgroup specificity, and genotype diversity of rotavirus strains in children with acute diarrhea in Paraguay. J. Clin. Microbiol. 40:1709-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coulson, B. S., and C. Kirkwood. 1991. Relation of VP7 amino acid sequence to monoclonal antibody neutralization of rotavirus and rotavirus monotype. J. Virol. 65:5968-5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyall-Smith, M. L., I. Lazdins, G. W. Tregear, and I. H. Holmes. 1986. Location of the major antigenic sites involved in rotavirus serotype-specific neutralization. Proc. Natl. Acad. Sci. USA 83:3465-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espinoza, F. 2004. Rotavirus in pediatric gastroenteritis in Nicaraguan children. Ph.D. dissertation. University of Léon, Léon, Nicaragua.
- 13.Espinoza, F., F. Bucardo, M. Paniagua, L. Svensson, H. O. Hallander, and K. Bondeson. 2006. Shifts of rotavirus G and P types in Nicaragua—2001-2003. Pediatr. Infect. Dis. J. 25:1078-1080. [DOI] [PubMed] [Google Scholar]
- 14.Espinoza, F., M. Paniagua, H. Hallander, L. Svensson, and O. Strannegard. 1997. Rotavirus infections in young Nicaraguan children. Pediatr. Infect. Dis. J. 16:564-571. [DOI] [PubMed] [Google Scholar]
- 15.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z. Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iturriza-Gomara, M., D. Cubitt, U. Desselberger, and J. Gray. 2001. Amino acid substitution within the VP7 protein of G2 rotavirus strains associated with failure to serotype. J. Clin. Microbiol. 39:3796-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iturriza-Gomara, M., U. Desselberger, and J. Gray. 2003. Molecular epidemiology of rotaviruses: genetic mechanisms associated with diversity, p. 317-344. In U. Desselberger and J. Gray (ed.), Viral gastroenteritis. Perspectives in medical virology, vol. 9. Elsevier Science B.V., Amsterdam, The Netherlands. [Google Scholar]
- 19.Iturriza-Gomara, M., J. Green, D. W. Brown, U. Desselberger, and J. J. Gray. 1999. Comparison of specific and random priming in the reverse transcriptase polymerase chain reaction for genotyping group A rotaviruses. J. Virol. Methods 78:93-103. [DOI] [PubMed] [Google Scholar]
- 20.Iturriza-Gomara, M., J. Green, D. W. Brown, M. Ramsay, U. Desselberger, and J. J. Gray. 2000. Molecular epidemiology of human group A rotavirus infections in the United Kingdom between 1995 and 1998. J. Clin. Microbiol. 38:4394-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iturriza-Gomara, M., G. Kang, and J. Gray. 2004. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J. Clin. Virol. 31:259-265. [DOI] [PubMed] [Google Scholar]
- 22.Kapikian, A. Z., Y. Hoshino, and, C. R. M. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 23.Kirkwood, C., N. Bogdanovic-Sakran, E. Palombo, P. Masendycz, H. Bugg, G. Barnes, and R. Bishop. 2003. Genetic and antigenic characterization of rotavirus serotype G9 strains isolated in Australia between 1997 and 2001. J. Clin. Microbiol. 41:3649-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kudo, S., Y. Zhou, X. R. Cao, S. Yamanishi, S. Nakata, and H. Ushijima. 2001. Molecular characterization in the VP7, VP4 and NSP4 genes of human rotavirus serotype 4 (G4) isolated in Japan and Kenya. Microbiol. Immunol. 45:167-171. [DOI] [PubMed] [Google Scholar]
- 25.Laird, A. R., J. R. Gentsch, T. Nakagomi, O. Nakagomi, and R. I. Glass. 2003. Characterization of serotype G9 rotavirus strains isolated in the United States and India from 1993 to 2001. J. Clin. Microbiol. 41:3100-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liakopoulou, E., K. Mutton, D. Carrington, S. Robinson, C. G. Steward, N. J. Goulden, J. M. Cornish, and D. I. Marks. 2005. Rotavirus as a significant cause of prolonged diarrhoeal illness and morbidity following allogeneic bone marrow transplantation. Bone Marrow Transplant 36:691-694. [DOI] [PubMed] [Google Scholar]
- 27.Ministerio de Salud de Nicaragua. 2005. Boletin Epidemiologico del Ministerio de Salud de Nicaragua, no. 13. Ministerio de Salud de Nicaragua, Managua, Nicaragua.
- 28.Nakajima, H., T. Nakagomi, T. Kamisawa, N. Sakaki, K. Muramoto, T. Mikami, H. Nara, and O. Nakagomi. 2001. Winter seasonality and rotavirus diarrhoea in adults. Lancet 357:1950. [DOI] [PubMed] [Google Scholar]
- 29.Nilsson, M., K. O. Hedlund, M. Thorhagen, G. Larson, K. Johansen, A. Ekspong, and L. Svensson. 2003. Evolution of human calicivirus RNA in vivo: accumulation of mutations in the protruding P2 domain of the capsid leads to structural changes and possibly a new phenotype. J. Virol. 77:13117-13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Mahony, J., B. Foley, S. Morgan, J. G. Morgan, and C. Hill. 1999. VP4 and VP7 genotyping of rotavirus samples recovered from infected children in Ireland over a 3-year period. J. Clin. Microbiol. 37:1699-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pager, C. T., J. J. Alexander, and A. D. Steele. 2000. South African G4P[6] asymptomatic and symptomatic neonatal rotavirus strains differ in their NSP4, VP8*, and VP7 genes. J. Med. Virol. 62:208-216. [DOI] [PubMed] [Google Scholar]
- 32.Parashar, U. D., E. G. Hummelman, J. S. Bresee, M. A. Miller, and R. I. Glass. 2003. Global illness and deaths caused by rotavirus disease in children. Emerg. Infect. Dis. 9:565-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubilar-Abreu, E., K. O. Hedlund, L. Svensson, and C. Mittelholzer. 2005. Serotype G9 rotavirus infections in adults in Sweden. J. Clin. Microbiol. 43:1374-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steele, D., E. Reynecke, M. de Beer, P. Bos, and I. Smuts. 2002. Characterization of rotavirus infection in a hospital neonatal unit in Pretoria, South Africa. J. Trop. Pediatr. 48:167-171. [DOI] [PubMed] [Google Scholar]
- 35.Svensson, L., I. Uhnoo, M. Grandien, and G. Wadell. 1986. Molecular epidemiology of rotavirus infections in Uppsala, Sweden, 1981: disappearance of a predominant electropherotype. J. Med. Virol. 18:101-111. [DOI] [PubMed] [Google Scholar]
- 36.Volotao, E. M., C. C. Soares, A. G. Maranhao, L. N. Rocha, Y. Hoshino, and N. Santos. 2006. Rotavirus surveillance in the city of Rio de Janeiro-Brazil during 2000-2004: detection of unusual strains with G8P[4] or G10P[9] specificities. J. Med. Virol. 78:263-272. [DOI] [PubMed] [Google Scholar]
- 37.Zao, C. L., W. N. Yu, C. L. Kao, K. Taniguchi, C. Y. Lee, and C. N. Lee. 1999. Sequence analysis of VP1 and VP7 genes suggests occurrence of a reassortant of G2 rotavirus responsible for an epidemic of gastroenteritis. J. Gen. Virol. 80:1407-1415. [DOI] [PubMed] [Google Scholar]