The late Richard Feynman once said, “What I cannot create, I do not understand.” Although this principle is well known to physicists and engineers, it has only recently become the mantra of a cadre of scientists seeking to build new biological pathways in living animals. In this issue of PNAS, Armbruster et al. (1) report an important new tool for pharmacologists seeking to build designer G protein-coupled receptors (GPCRs) to control signaling pathways. This family of receptors has been fine-tuned over millions of years to sense molecules ranging from yeast pheromones to neurotransmitters. Nature has evolved hundreds of human GPCRs, and the pharmaceutical industry spends billions of dollars to develop GPCR drugs that frequently top the list of best-selling medicines. However, despite intense studies, many mysteries remain about how GPCRs function in vivo. Although GPCR drugs and gene knockouts have taught us a great deal, some lessons may only be learned by building new GPCR signaling pathways in living animals.
The first attempt at making a designer GPCR was led by Catherine Strader (2), who developed a series of compounds to selectively activate a mutant version of the β-adrenergic receptor that was unresponsive to its natural hormone. Unfortunately, this elegant work only yielded compounds with millimolar affinities, with unknown pharmacokinetics, making in vivo work impractical.
Later, my laboratory devised a series of RASSLs (receptors activated solely by a synthetic ligand) with nanomolar agonists, making in vivo use possible for the first time (3). The key to making these first RASSLs was to take advantage of potent preexisting synthetic drugs, such as kappa opioid agonists (e.g., spiradoline), that had been developed by the pharmaceutical industry as potential analgesics. A RASSL could be made by introducing mutations that abrogated signaling via the natural peptide ligands yet preserved stimulation via the drug, spiradoline. The first in this series, Ro1 (RASSL opioid no. 1), has been expressed in at least six different tissues in transgenic animals (4) to induce diverse phenotypes, such as heart-rate control (5), cardiomyopathy (4), and taste sensation (6, 7).
Once scientists realized that RASSLs could be made by using existing drugs, RASSLs soon emerged from studies using a wide variety of receptors, including the 5HT4 (8), α2-adrenergic (9), histamine (10), and melanocortin (11) receptors. These initial in vivo experiments have been filled with surprises and whet our appetite for more RASSLs with improved ligands and a wider range of signaling responses. Using preexisting drugs allows scientists to take advantage of known pharmacology, and in several cases (e.g., 5HT4, agonists), the side effects are negligible in animals. Furthermore, recent studies with RASSLs derived from the 5HT4 (8) and histamine (10) receptors have identified drugs that are far more potent in activating the RASSL than the endogenous receptor. Still, these first- generation RASSLs cannot have a “clean” background for experimentation unless the endogenous receptor gene is knocked out, requiring complex genetic crosses (12).
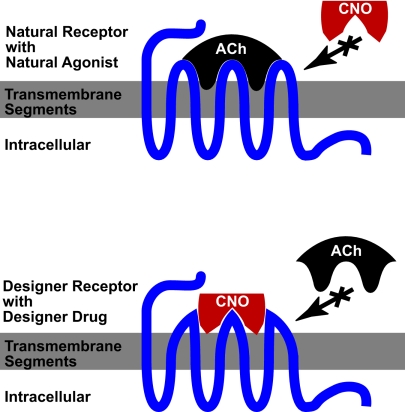
Now, Armbruster et al. (1) have developed an elegant solution to this problem, using the powerful tools of directed evolution. They use a well established yeast mutagenesis system to produce thousands of mutant hM3 muscarinic receptors, which they screened for signaling characteristics of an ideal RASSL. In a tour-de-force of molecular pharmacology, they use multiple rounds of mutagenesis and repeat screening to isolate mutants that had lost the ability to respond to the natural ligand (acetylcholine) but gained the ability to respond to the inert compound, clozapine-N-oxide (CNO) with nanomolar potencies (see Fig. 1).
Fig. 1.
Construction of a designer receptor. Native hM3 receptor responds to the natural hormone (Ach) but does not respond to CNO (Upper). This receptor is matured through saturation mutagenesis to result in a designer receptor (Lower), which responds only to the designer drug (CNO) and is impervious to the natural hormone.
Armbruster et al. (1) dubbed this new class of receptors as DREADDs (designer receptors exclusively activated by a designer drug) with the first in the series being named hM3D. The hM3 muscarinic receptor activates the Gq pathway to cause calcium mobilization, whereas its close relatives (M2 and M4) inhibit cAMP accumulation via the Gi pathway. By making analogous mutations in these receptors, they produce hM2D and hM4D that now activate the Gi pathway in response to CNO. Thus, CNO could activate either the Gq or the Gi signaling pathways, depending on which RASSL is expressed. Armbruster et al. go on to demonstrate these signaling properties in tissue culture cells and transfected hippocampal slices.
The real test for this new class of designer receptor still lies in its in vivo use because the pharmacology of CNO is relatively unexplored. However, there is great promise because CNO is thought to be biologically inert and has undergone limited human studies (1). If CNO is biologically inert, it could provide a “blank slate” on which scientists can express and activate RASSLs without concern for other confounding effects of the drug. Furthermore, a family of RASSLs activated by the same drug that each have different biochemical signals will allow scientists to directly compare each receptor response while controlling for pharmacokinetics of the drug. Finally, Armbruster et al. (1) carefully screened for receptors with low basal activity that could provide a maximal dynamic range in agonist-induced responses.
The use of directed evolution to design new GPCRs is a true milestone in our quest to build synthetic GPCR signaling pathways, although many challenges still lie ahead. Once a RASSL is expressed, it has an active “social life,” cycling to and from the plasma membrane, coupling to G proteins, other receptors, and a wide variety of other intracellular molecules (13). Currently, the in vivo function of most signaling molecules remains unclear because we are limited to knockout and overexpression studies. One can now envision an additional series of RASSLs that have been engineered to tether to different scaffolding proteins, ion channels, or intracellular signaling molecules. Ligand activation could provide temporal control of these molecules that otherwise would be difficult to engineer. For example, it is clear that Gs-coupled receptors aid in memory formation, speed heart rate, and relax smooth muscles via increases in cAMP, yet it is unclear how these responses are modulated by other intracellular signaling molecules. Optimal control of a physiological response may require GPCRs to directly couple to a specific arrestin, ion channel, or kinase. An orthogonal RASSL signaling system may provide a new way to answer these intriguing questions.
Another major challenge will be to control when, where, and to what extent a RASSL is expressed. We are learning that basal activity is an essential component for the normal function of many GPCRs (13). We may need to engineer RASSLs that have various levels of basal activity to truly recapitulate a native receptor. Even receptors with no measurable basal biochemical response in tissue culture can exhibit basal physiological responses when expressed at high levels in animals (4). Because RASSLs have such potent physiological effects, we have found they are best studied by using conditional expression systems, such as the tetracycline transactivator (Tet) system (14). With the Tet system, a single RASSL transgenic line can be used to drive expression in multiple tissues with tight temporal control. Using these tools, one can imagine directing RASSLs to rewire and dissect complex physiological processes, such as memory, addiction, or organ development.
Finally, some groups have suggested that RASSLs could be used to selectively deliver GPCR responses for therapeutic purposes (3, 8). For example, in Parkinson's disease and seizure disorders, hyperactive neural activity could be modulated by RASSLs expressed at specific locations. Similar scenarios could be imagined for controlling growth or differentiation of embryonic stem cells. Although any therapeutic use of RASSLs is certainly many years away, this process would be accelerated by inert agonists that are already approved for human use. For instance, many antibiotics have similar properties to GPCR drugs, and some are even weak agonists of endogenous GPCRs (motilin receptor) (15). It is conceivable that directed evolution could be used to create a RASSL that responds to an antibiotic or other inert molecule already approved for human use. Although these “old” molecules could hardly be considered “designer drugs,” their well known pharmacokinetics and safety record could provide ideal RASSL agonists.
Ligand activation could provide temporal control of molecules that otherwise would be difficult to engineer.
The primary benefit of synthetic GPCR systems will most likely be an increased understanding of how natural GPCRs work. For instance, if we could use RASSLs to identify GPCR signals that induce embryonic stem cells to develop into pancreatic islet cells, this would help focus a search for drugs acting on endogenous GPCRs to have the same effect. Similarly, if RASSLs could help implicate new effector molecules that orchestrate heart-rate control, this could reveal new therapeutic targets for cardiac arrhythmias. Because GPCRs are such ideal drug targets, any new insight into this system is likely to lead to new therapeutic approaches to modulate the endogenous receptor systems.
Although synthetic biology is still in its infancy, we can already see its benefits. Building the first synthetic GPCR signaling pathways with RASSLs has already provided many lessons and surprises. Armbruster et al. (1) may refer to this new class of RASSLs as DREADDs, but the field of synthetic biology is celebrating, rather than dreading, these powerful new additions to our toolbox. Many of the most perplexing biological questions today involve complex tissues, and the availability of a synthetic signaling system activated by an inert drug provides great experimental opportunities. Now, perhaps by following Richard Feynman's advice, we can continue to build so that we can understand.
Footnotes
The author declares no conflict of interest.
See companion article on page 5163.
References
- 1.Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Proc Natl Acad Sci USA. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strader C, Gaffney T, Sugg E, Candelore M, Keys R, Patchett A, Dixon R. J Biol Chem. 1991;266:5–8. [PubMed] [Google Scholar]
- 3.Coward P, Wada H, Falk M, Chan S, Meng F, Akil H, Conklin B. Proc Natl Acad Sci USA. 1998;95:352–357. doi: 10.1073/pnas.95.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scearce-Levie K, Coward P, Redfern C, Conklin B. Trends Pharmacol Sci. 2001;22:414–420. doi: 10.1016/s0165-6147(00)01743-0. [DOI] [PubMed] [Google Scholar]
- 5.Redfern C, Coward P, Degtyarev M, Lee E, Kwa A, Hennighausen L, Bujard H, Fishman G, Conklin BR. Nat Biotechnol. 1999;17:165–169. doi: 10.1038/6165. [DOI] [PubMed] [Google Scholar]
- 6.Mueller K, Hoon M, Erlenbach I, Chandrashekar J, Zuker C, Ryba N. Nature. 2005;434:225–229. doi: 10.1038/nature03352. [DOI] [PubMed] [Google Scholar]
- 7.Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJ, Zuker CS. Cell. 2003;115:255–266. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]
- 8.Claeysen S, Joubert L, Sebben M, Bockaert J, Dumuis A. J Biol Chem. 2003;278:699–702. doi: 10.1074/jbc.C200588200. [DOI] [PubMed] [Google Scholar]
- 9.Pauwels P. Trends Pharm Sci. 2003;24:504–507. doi: 10.1016/j.tips.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Bruysters M, Jongejan A, Akdemir A, Bakker R, Leurs R. J Biol Chem. 2005;280:34741–34746. doi: 10.1074/jbc.M504165200. [DOI] [PubMed] [Google Scholar]
- 11.Srinivasan S, Vaisse C, Conklin B. Ann NY Acad Sci. 2003;994:225–232. doi: 10.1111/j.1749-6632.2003.tb03184.x. [DOI] [PubMed] [Google Scholar]
- 12.Sweger EJ, Casper KB, Scearce-Levie K, Conklin BR, McCarthy KD. J Neurosci. 2007;27:2309–2317. doi: 10.1523/JNEUROSCI.4565-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spiegel A, Weinstein L. Annu Rev Med. 2004;55:27–39. doi: 10.1146/annurev.med.55.091902.103843. [DOI] [PubMed] [Google Scholar]
- 14.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 15.Feighner S, Tan C, McKee K, Palyha O, Hreniuk D, Pong S, Austin C, Figueroa D, MacNeil D, Cascieri M, et al. Science. 1999;284:2184–2188. doi: 10.1126/science.284.5423.2184. [DOI] [PubMed] [Google Scholar]