Abstract
The mechanism that nature applies to dissipate excess energy from solar UV light absorption in DNA is fundamental, because its efficiency determines the vulnerability of all genetic material to photodamage and subsequent mutations. Using femtosecond time-resolved broadband spectroscopy, we have traced the electronic excitation in both time and space along the base stack in a series of single-stranded and double-stranded DNA oligonucleotides. The obtained results demonstrate not only the presence of delocalized electronic domains (excitons) as a result of UV light absorption, but also reveal the spatial extent of the excitons.
Keywords: excitons, femtosecond, spectroscopy, photophysics
Since the early days of DNA photochemistry there has been considerable speculation about the nature of the excited singlet states of the bases, i.e., whether they are localized on a single base or delocalized over several bases (1–4). The answer to this question is paramount for understanding the mechanism of UV light-induced chemical reactions that may result in carcinogenic mutations (5–7). Although the rich photochemistry of nucleic acids has been extensively studied (5, 6), the precursor states for chemical reactions damaging the genome are not well characterized. In other words, the nature of these states and the concomitant electronic and nuclear dynamics must be investigated to understand how excitation energy is transformed and dissipated within the double-helix. With respect to the biological consequences of UV light-induced phenomena in DNA most efforts were spent on the investigation of charge migration processes through the base stack (8, 9). Although photoinduced charge transfer is initiated by electronic excitation, no clear picture has yet emerged of how to describe electronic excitations in DNA. Circular dichroism measurements on single-stranded homoadenines and A·T duplexes indicate significant electronic coupling between the adjacent bases in the stack (10). On the other hand, the fact that the DNA UV spectra closely resemble the sum of the spectra of the constituent bases was interpreted as evidence for strictly localized excited states (1). Since the pioneering work of Eisinger et al. (2), who reported a broad red-shifted emission in high-concentrated nucleoside solutions at low temperatures, excimers and exciplexes have been invoked in discussions on nucleic acid (11–17). What remains unclear, however, is the question of whether these states are formed as a result of dynamic quenching, i.e., through combined nuclear rearrangements of locally excited bases with nonexcited adjacent bases. Alternatively, the UV excitation of DNA could have excitonic character with shared oscillator strength between adjacent bases. In this article, we provide firm experimental results that indicate the presence of electronically delocalized domains in DNA base stacks as a result of UV light absorption. Our femtosecond spectroscopic data are interpreted and discussed in the framework of molecular exciton theory (18). The first theoretical studies on molecular excitons in DNA were reported ≈40 years ago (19). Since then, more sophisticated calculations combining quantum chemistry and molecular dynamics have followed (17, 20, 21). Emanuele et al. (21) recently predicted that the excitation is delocalized over at least two bases.
Several years ago, we (16) reported spectroscopic evidence for electronic delocalization in double-stranded oligonucleotides that contained pairs of a fluorescent DNA base analog. In the present study, we investigated natural single-stranded DNA (dA)n [composed of n = 2, 3, 4, 5, 6, 12, 15, 18 consecutive 2′-deoxyadenosine (dA) residues, respectively], as well as double-stranded oligonucleotides (dA)n·(dT)n (n = 12, 18) and (dAT)9·(dAT)9. The single-stranded homoadenine oligonucleotides were chosen because of their reported helical stack structures in aqueous solutions and the option to systematically control the length of the stack. NMR studies and spectroscopic titrations (22) on (dA)2 have revealed that 80% of the dinucleotides form a B-DNA helical structure at room temperature, whereas for (dA)3 the number is 86% (23). Furthermore, Kohler and coworkers (24) have recently shown that vertical stacking interactions, and not base pairing, dominate the fate of the excited base states.
Results and Discussion
Femtosecond broadband pump-probe spectroscopy enables us to monitor the temporal evolution of the excited-state absorption (ESA), including the absorption of “dark states” (i.e., states with negligible fluorescence quantum yields), which are dominant in excited DNA. The combination of broadband spectral probing (from 300 to 700 nm) with a time resolution of <100 fs allows us to monitor a variety of spectroscopic transitions simultaneously. Some of these transitions are local, i.e., their participating wave functions are localized on individual chromophores or molecular residues. These local transitions are often not very sensitive to changes in the molecular structure that lead to alterations of the interaction between chromophores. On the other hand, there are transitions composed of wavefunctions which are delocalized between different chromophores. Those interchromophoric transitions are generally very sensitive to changes in the molecular structure and electronic delocalization (25, 26).
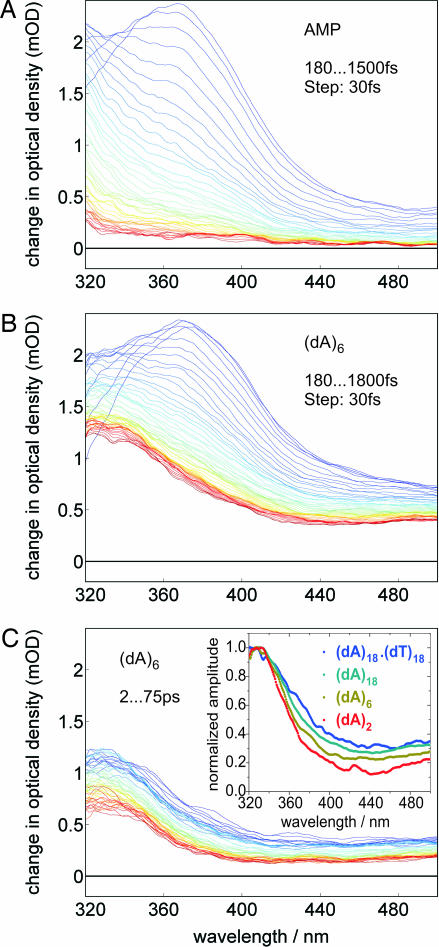
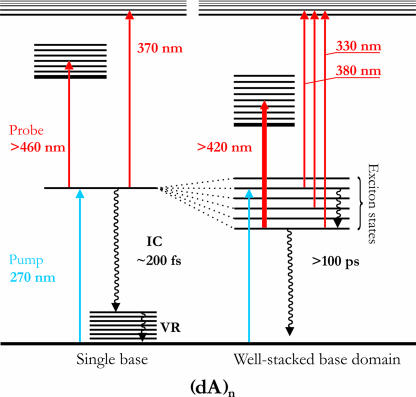
Within the first 100 fs after optical excitation the ESA spectra of all nucleic acids studied here change drastically. As reported (27–29), the initially excited state of AMP decays to a vibrationally hot ground state with a time constant of <200 fs. In our experiment, the hot ground-state absorption manifests itself as a broad spectral tail (30) with monotonically rising intensity toward the short-wavelength probing limit at 300 nm (Fig. 1A). Fig. 1B displays the early-time spectral dynamics of (dA)6 where the initial band at ≈380 nm decays and a new band rises at ≈330 nm. (dA)6 was chosen representatively for all (dA)n systems that show qualitatively identical spectral dynamics. The time scale for the formation of the ESA band ≈330 nm is sub-1 ps in all (dA)n systems studied. Dynamic quenching, i.e., the shortening of this ultrafast time component in (dA)n oligomers (with respect to AMP) caused by excimer formation, has not been observed. After 1.5 ps the spectral position of the maximum of the ESA spectrum remains constant (at 330 nm) throughout the excited-state lifetime of several 100 ps (Fig. 1C). The observed spectral dynamics can be illustrated in a simple energy level diagram (Fig. 2). Depending on the local environment of each adenine, one may simplistically distinguish two categories of chromophores: (i) well stacked base domains and (ii) poorly stacked single bases. For well stacked base domains, UV absorption leads to delocalized exciton states where the electronic wave functions are spread over more than a single base. After optical excitation the initially excited states undergo electronic relaxation within the exciton-state manifold, characterized by a pronounced spectral blue shift (from 380 to 330 nm). Entirely consistent with this internal conversion (IC) process is the observed change (i.e., decrease) in oscillator strength. The ultrafast electronic relaxation (sub-100 fs, in agreement with ref. 31) is followed by a slower nuclear relaxation process, which may involve modes from the base stack and from the surrounding medium and manifests itself in minor changes of the 330-nm band shape. A comparison of the short-wavelength ESA bands (<380 nm) in AMP and (dA)n indicates that the underlying electronic transition is localized and thus insensitive to electronic delocalization. On the other hand, the broad ESA band >420 nm results from interchromophoric interactions and reflects electronic delocalization in the system (see below). The electronically relaxed exciton state has a lifetime of several 100 ps, in accordance with results from previously reported transient absorption measurements (24, 32). For poorly stacked single bases, parallel to the optical excitation of vertically stacked base domains, monomer-like excitations of single adenine bases occur at places in the sequence where local static and dynamic disorder disrupts the electronic coupling between neighboring bases. These localized adenine states resemble similarity with AMP, which undergoes sub-200-fs IC to its vibrationally hot ground state. The typical spectral fingerprint of hot ground states in all (dA)n systems (as seen in AMP; Fig. 1A) is superimposed to the 330- to 380-nm ESA bands.
Fig. 1.
Temporal evolution of the ESA spectra of AMP (A) and (dA)6 (B and C). Early spectra are shown in blue/green, and late spectra are in orange/red. (C Inset) The averaged normalized spectra between 5 and 9 ps of (dA)2, (dA)6, (dA)18, and (dA)18·(dT)18. All spectra shown have been corrected for solvent contributions (see Materials and Methods for details).
Fig. 2.
Energy-level diagram for the energy dissipation pathways in homoadenine sequences (dA)n. In addition to poorly stacked single bases that show very similar behavior as AMP, there are well stacked base domains that can be excited cooperatively by UV light, forming delocalized exciton states. Energy dissipation in these delocalized states involves IC and can be traced spectroscopically (see Results and Discussion for details).
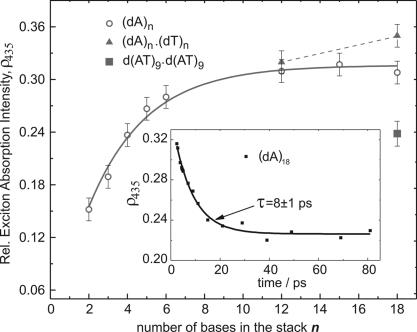
Hence, there are two excited-state processes that occur simultaneously in (dA)n oligonucleotides: (i) a sub-200-fs IC from the initially excited state forming the hot ground state (similar to AMP) and (ii) electronic relaxation (IC) of delocalized exciton states. Fig. 1C Inset displays the ESA spectra of (dA)2, (dA)6, and (dA)18 5 ps after excitation. Clearly, all spectra contain the same 330-nm ESA band. However, the long-wavelength part (>420 nm) of the spectrum depends on the length of the base stack n. This critical observation indicates that the electronic composition of the excited-base states varies with the number of individual bases in the stack and is a manifestation for exciton delocalization. The intensity ratio ρ435, between the long-wavelength part of the ESA spectrum, dominated by interchromophoric transitions centered at ≈435 nm, and the local transition at 330 nm, is proportional to the oscillator strength for exciton absorption. Thus measuring ρ435 as a function of n will provide information about the spatial extent of the exciton.† For instance, if the excitation were only spread over two adjacent bases, ρ435 would not change with increasing length of the stack because the effective number of stacked adenine dimers is approximately constant in all samples. On the other hand, ρ435 would continuously increase with increasing n if the excitation were completely delocalized over all bases in the stack. Fig. 3 shows ρ435 after 3 ps for all (dA)n systems and for three A·T DNA duplexes. Two observations are critical: (i) ρ435 increases monotonically with increasing length of the base stack, and (ii) ρ435 is not constant over the excited-state lifetime of the bases but decays with a characteristic time constant of 8–10 ps in all DNA systems studied, except for (dAT)9·(dAT)9 oligonucleotides (see Fig. 3 Inset). Structural conversions in nucleic acids such as local transitions from A- to B-DNA are known to occur on time scales several orders of magnitude slower than the observed 10 ps (33). Therefore, one must conclude that changes in the relative spectral intensities during the excited-state lifetime of the bases are caused by small-amplitude fluctuations in the base stack and/or the surrounding environment (metal ions, solvent) (34).
Fig. 3.
Spectral intensity ratio between the exciton absorption (in a 30-nm interval, centered at 435 nm) and the monomer absorption at 330 nm (ρ435), 3 ps after excitation, as a function of the stack length n. For the (dA)n series, a single exponential fit (solid curve) was used to extract the “1/e delocalization length” d. The ρ435 values for the A·T duplexes are connected with a dotted line. (Inset) Time dependence of ρ435 for (dA)18. The decay of ρ435 is characterized by time constants of 8–10 ps in all DNA systems studied, except for (dAT)9·(dAT)9 where no decay of ρ435 was observed (see Results and Discussion for details).
Fig. 3 shows larger changes of ρ435(n) at short stack lengths and a more gradual evolution for larger n. The obtained base stack dependence of ρ435 reflects an exponential decay of the exciton delocalization length in DNA and reveals a “1/e delocalization length” d for (dA)n of 3.3 ± 0.5 bp. Because shorter duplexes are not thermodynamically stable at room temperature, the delocalization length in A·T duplexes can only be estimated by comparing (dA)12·(dT)12 and (dA)18·(dT)18. Although ρ435 has not reached saturation at n = 12 and continues to increase toward n = 18, the relative change is small compared with the increase of ρ435 at shorter stack lengths [i.e., between (dA)2 and (dA)5)]. Given the structural similarities with respect to stacking in single-stranded homoadenine sequences and A·T duplexes, these results suggest a similar length distribution of the delocalized domains in both types of nucleic acids, albeit A·T duplexes appear to have a slightly larger fraction of more extended (d > 4) delocalized domains. The overall larger values of ρ435 reflect stronger exciton absorption and thus larger electronic coupling because of shorter base–base distances and/or a more rigid stack structure in duplexes as opposed to single-stranded sequences.
It is interesting to note that the alternating sequence (dAT)9·(dAT)9 reveals an intermediate ρ435 value that corresponds to the degree of delocalization found in (dA)4. In contrast to all of the other nucleic acid systems with vertically stacked adenines (in the same strand), ρ435 in (dAT)9·(dAT)9 does not change with time. This observation may simply reflect the different electronic character of intrastrand excited A-T-A… complexes, compared with A-A-A… domains. For instance, one would expect the former complexes to have substantial charge transfer contributions that would not participate in the exciton state wavefunctions formed in homadenines.
Conclusions
Guided by the temporal evolution of the ESA spectra shown in Fig. 1, the following conclusions emerge. The electronic coupling between stacked bases leads to the formation of delocalized exciton states upon UV absorption. In single-stranded homoadenine sequences the typical 1/e delocalization length is 3–4 bases. However, given the conformational inhomogeneiety of these flexible biopolymers, more extended delocalization is likely to be present in some molecules. Ensembles of A·T duplexes have a larger fraction of more extended delocalized domains. This is evidenced by the significant increase in ρ435 from (dA)12·(dT)12 to (dA)18·(dT)18. The observed temporal decay of ρ435 is a clear indication that the electronic exciton structure is dynamic, and that changes in the delocalization length occur during the lifetime of the excited base stack. Because of the observed time scale, these changes must be induced by nuclear motions within the base stack (35) and/or in the immediate surroundings of the DNA. Finally, our results have shown that, despite favorable stacking and apparent strong electronic coupling, there is a substantial fraction of excited DNA molecules that undergo ultrafast IC to the hot ground state, similar as in single bases (24, 32). It is reasonable to assume that the optical excitation in this molecular subensemble remains localized due to static and dynamic disorder in the stack. In fact, random DNA sequences containing both A·T and G·C base pairs are even more likely to yield localized “monomer-type” electronic states. The effective competition of the monomer-type photophysical pathway with the exciton formation is critical from an evolutionary viewpoint because excess energy is funneled to the ground state in times shorter than needed to make and break chemical bonds. The amount of disorder in the genome will define what fraction of stacked bases can avoid irreversible photo damage by eliminating electronic excess energy in the same fashion as single DNA bases.
Materials and Methods
Femtosecond Broadband Pump-Probe Measurement.
The experimental setup has been described in detail (26). Briefly, for generating the pump and probe pulses a commercial CPA-2010 (775 nm, 150 fs, 1 mJ, 1 kHz) from Clark-MXR (Dexter, MI) was used. Part of the laser output created the 270-nm pump beam using the frequency doubled output of a home-built noncollinear optical parametric amplifier system as the light converter. A small fraction of the 775-nm beam was used to generate the broadband white light continuum between 300 and 750 nm. Artifacts from the solvent were carefully subtracted from the ESA signal following the procedure by Lorenc et al. (36).
Sample Preparation.
AMP was obtained from Sigma/Aldrich (St. Louis, MO) and used without further purification. All RP-HPLC-purified oligodeoxynucleotides were purchased from Midland Certified Reagent Company (Midland, TX). All samples were prepared in 0.25 M NaCl and 25 mM sodium phosphate buffer (pH 7.15). The double-stranded oligomers (dA)12·(dT)12 and (dA)18·(dT)18 were prepared by mixing equimolar amounts of respective single strands and thermally annealed. All double-stranded oligomers, (dA)12·(dT)12, (dA)18·(dT)18, and d(AT)9·d(AT)9, were heated to 80°C for 10 min, cooled slowly down to room temperature, and used for pump-probe measurement immediately. All sample concentrations were adjusted to have similar absorbance of ≈0.9 at 270 nm, and pump-probe experiments were conducted at 22°C.
Acknowledgments
We thank Dimitar Popminchev, Stoichko Dimitrov, and Aleksandra Gotsova for valuable assistance in the data analysis process. This work was supported by National Science Foundation Grant CHE 0628119 and Boston College.
Abbreviations
- dA
deoxyadenosine
- ESA
excited-state absorption
- IC
internal conversion.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
A similar analysis to determine the exciton delocalization length in bacterial light-harvesting antenna systems was applied in ref. 37.
References
- 1.Eisinger J, Shulman RG. Science. 1968;161:1311–1319. doi: 10.1126/science.161.3848.1311. [DOI] [PubMed] [Google Scholar]
- 2.Eisinger J, Gueron M, Shulman RG, Yamane T. Proc Natl Acad Sci USA. 1966;55:1015–1020. doi: 10.1073/pnas.55.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gueron M, Shulman RG, Eisinger J. Proc Natl Acad Sci USA. 1966;56:814–818. doi: 10.1073/pnas.56.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gueron M, Eisinger J, Lamola AA. In: Basic Principles in Nucleic Acid Chemistry. Ts'O POP, Eisinger J, editors. New York: Academic; 1974. pp. 311–398. [Google Scholar]
- 5.Cadet J, Vigny P. In: Bioorganic Photochemistry. Morrison H, editor. Vol 1. New York: Wiley; 1990. pp. 1–272. [Google Scholar]
- 6.Ravanat JL, Douki T, Cadet J. J Photochem Photobiol B. 2001;63:88–102. doi: 10.1016/s1011-1344(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 7.Dumaz N, VanKranen HJ, DeVries A, Berg RJW, Wester PW, VanKreijl CF, Sarasin A, Daya-Grosjean L, DeGruijl FR. Carcinogenesis. 1997;18:897–904. doi: 10.1093/carcin/18.5.897. [DOI] [PubMed] [Google Scholar]
- 8.Wagenknecht HA, editor. Charge Transfer in DNA. Weinheim, Germany: Wiley; 2005. [Google Scholar]
- 9.Kaden P, Mayer-Enthart E, Trifonov A, Fiebig T, Wagenknecht H-A. Angew Chem Int Ed. 2005;44:1636–1639. doi: 10.1002/anie.200462592. [DOI] [PubMed] [Google Scholar]
- 10.Johnson WC, Tinoco I., Jr Biopolymers. 1969;7:727–749. [Google Scholar]
- 11.Garestier TM, Helene C, Michelson AM. Biochim Biophys Acta. 1969;182:342–354. [PubMed] [Google Scholar]
- 12.Morgan JP, Daniels M. Photochem Photobiol. 1980;31:207–213. [Google Scholar]
- 13.Ballini JP, Vigny P, Daniels M. Biophys Chem. 1983;18:61–65. doi: 10.1016/0301-4622(83)80027-1. [DOI] [PubMed] [Google Scholar]
- 14.Georghiou S, Zhu S, Weidner R, Huang C-R, Ge G. J Biomol Struct Dyn. 1990;8:657–674. doi: 10.1080/07391102.1990.10507834. [DOI] [PubMed] [Google Scholar]
- 15.Henderson PT, Boone E, Schuster GB. Helv Chim Acta. 2002;85:135–151. [Google Scholar]
- 16.Rist M, Wagenknecht H-A, Fiebig T. ChemPhysChem. 2002;3:704–707. doi: 10.1002/1439-7641(20020816)3:8<704::AID-CPHC704>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 17.Bouvier B, Gustavsson T, Markovitsi D, Millié P. Chem Phys. 2002;275:75–92. [Google Scholar]
- 18.Davydov AS. Theory of Molecular Excitons. New York: McGraw–Hill; 1962. [Google Scholar]
- 19.Tinoco I., Jr J Am Chem Soc. 1960;82:4785–4790. [Google Scholar]
- 20.Bouvier B, Dognon JP, Lavery R, Markovitsi D, Millie P, Onidas D, Zakrzewska K. J Phys Chem B. 2003;107:13512–13522. [Google Scholar]
- 21.Emanuele E, Markovitsi D, Milli P, Zakrzewska K. ChemPhysChem. 2005;6:1387–1392. doi: 10.1002/cphc.200500014. [DOI] [PubMed] [Google Scholar]
- 22.Simpkins H, Richards EG. Biochemistry. 1967;6:2513–2520. doi: 10.1021/bi00860a031. [DOI] [PubMed] [Google Scholar]
- 23.Olsthoorn CSM, Bostelaar LJ, De Rooij JFM, Van Boom JH, Altona C. Eur J Biochem. 1981;115:309–321. doi: 10.1111/j.1432-1033.1981.tb05240.x. [DOI] [PubMed] [Google Scholar]
- 24.Crespo-Hernandez CE, Cohen B, Kohler B. Nature. 2005;436:1141–1144. doi: 10.1038/nature03933. [DOI] [PubMed] [Google Scholar]
- 25.Mank D, Raytchev M, Amthor S, Lambert C, Fiebig T. Chem Phys Lett. 2003;376:201–206. [Google Scholar]
- 26.Raytchev M, Pandurski E, Buchvarov I, Modrakowski C, Fiebig T. J Phys Chem A. 2003;107:4592–4600. [Google Scholar]
- 27.Peon J, Zewail AH. Chem Phys Lett. 2001;348:255–262. [Google Scholar]
- 28.Pecourt J-ML, Peon J, Kohler B. J Am Chem Soc. 2001;123:10370–10378. doi: 10.1021/ja0161453. [DOI] [PubMed] [Google Scholar]
- 29.Markovitsi D, Sharonov A, Onidas D, Gustavsson T. ChemPhysChem. 2003;4:303–305. doi: 10.1002/cphc.200390050. [DOI] [PubMed] [Google Scholar]
- 30.Sension RJ, Repinec ST, Hochstrasser RM. J Chem Phys. 1990;93:9185–9188. [Google Scholar]
- 31.Markovitsi D, Onidas D, Gustavsson T, Talbot F, Lazzarotto E. J Am Chem Soc. 2005;127:17130–17131. doi: 10.1021/ja054955z. [DOI] [PubMed] [Google Scholar]
- 32.Crespo-Hernandez CE, Kohler B. J Phys Chem B. 2004;108:11182–11188. [Google Scholar]
- 33.Bloomfield VA, Crothers DM, Tinoco I., Jr . Nucleic Acids: Structures, Properties, and Functions. Sausolito, CA: University Science Books; 2000. [Google Scholar]
- 34.Barnett RN, Cleveland CL, Joy A, Landman U, Schuster GB. Science. 2001;294:567–571. doi: 10.1126/science.1062864. [DOI] [PubMed] [Google Scholar]
- 35.Valis L, Wang Q, Raytchev M, Buchvarov I, Wagenknecht H-A, Fiebig T. Proc Natl Acad Sci USA. 2006;103:10192–10195. doi: 10.1073/pnas.0600957103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorenc M, Ziolek M, Naskrecki R, Karolczak J, Kubicki J, Maciejewski A. Appl Phys B. 2002;74:19–27. [Google Scholar]
- 37.Novoderezhkin V, Monshouwer R, van Grondelle R. Biophys J. 1999;77:666–681. doi: 10.1016/S0006-3495(99)76922-5. [DOI] [PMC free article] [PubMed] [Google Scholar]