Abstract
Human settlement of Oceania marked the culmination of a global colonization process that began when humans first left Africa at least 90,000 years ago. The precise origins and dispersal routes of the Austronesian peoples and the associated Lapita culture remain contentious, and numerous disparate models of dispersal (based primarily on linguistic, genetic, and archeological data) have been proposed. Here, through the use of mtDNA from 781 modern and ancient Sus specimens, we provide evidence for an early human-mediated translocation of the Sulawesi warty pig (Sus celebensis) to Flores and Timor and two later separate human-mediated dispersals of domestic pig (Sus scrofa) through Island Southeast Asia into Oceania. Of the later dispersal routes, one is unequivocally associated with the Neolithic (Lapita) and later Polynesian migrations and links modern and archeological Javan, Sumatran, Wallacean, and Oceanic pigs with mainland Southeast Asian S. scrofa. Archeological and genetic evidence shows these pigs were certainly introduced to islands east of the Wallace Line, including New Guinea, and that so-called “wild” pigs within this region are most likely feral descendants of domestic pigs introduced by early agriculturalists. The other later pig dispersal links mainland East Asian pigs to western Micronesia, Taiwan, and the Philippines. These results provide important data with which to test current models for human dispersal in the region.
Keywords: domestication, mtDNA, Pacific colonization, phylogeography
The peopling of Oceania was one of the most extensive human dispersals of the Holocene (1). Uncertainties remain, however, regarding the geographic origins of modern populations in Melanesia, Micronesia, and Polynesia and the origins of their ancestral cultures. A variety of scenarios have been inferred from associated material culture, language, and human genetic signatures to explain the movement of Neolithic cultures into Near and Remote Oceania (2–9). The degree to which these cultural and biological elements reflect dispersal has been questioned, as has the extent to which these various components were dispersed as a single unit (7). For example, models of the origins of Lapita (the immediate ancestors of the Polynesians and many other Oceanic cultures) that focus on the entire Lapita cultural and ecological package moving from Taiwan to the Pacific with little interaction (e.g., the “Express Train” or “Speedboat out of Taiwan”) are contrasted by others that identify broader regions and possibly multiple origins of the various cultural components.
Biological data can contribute to this debate through analyses of genetic variation in the domestic and commensal animals that were intimately linked with Neolithic cultures and were significant components of human dispersal and exchange networks. Pigs, chickens, dogs, and rats were introduced to the various islands of Near and Remote Oceania by early human settlers, and studies of Pacific rats (10) and pigs (11, 12) have demonstrated their potential as proxies for reconstructing patterns of human dispersal into Oceania. A previous genetic study of pigs (11) revealed a unique Pacific Clade, which included animals from Halmahera (in the Moluccas), New Guinea, and several Pacific islands, including Hawaii and Vanuatu. Current archeological evidence suggests that pigs appear in New Guinea (3) and the Moluccas (13) as late as 3,500 years B.P., probably linked with the arrival of nonindigenous agriculturalists. The absence of Pacific Clade haplotypes in any wild or domestic pigs from mainland Asia, or Island Southeast Asia (ISEA) west of the Wallace Line (11) in the previous study meant the origin of the clade could not be identified.
To investigate the geographic origin of the Pacific Clade and its distribution within ISEA and Near and Remote Oceania, we sequenced 663 bp of the mitochondrial control region from 243 wild, feral, and domestic pigs from across the region and mainland Southeast Asia, using primarily museum specimens. The modern specimens included: 160 from Malaysia and the Greater Sunda (including Sumatra, Java, Sulawesi, and numerous smaller adjacent islands), the Lesser Sunda chain (Bali, Lombok, Sumbawa, Sumba, Flores, and Timor), and other islands (including the Moluccas); 57 from the Philippines; and 9 from Polynesian islands. The 663-bp fragment was also determined for three archeological pig samples, and an additional 23 ancient samples were characterized with three diagnostic ≈120-bp fragments by using appropriate ancient DNA methods including independent replication (14, 15). An additional 512 pig sequences from GenBank (generating a total of 755) were used in phylogenetic analyses performed with Bayesian Monte Carlo–Markov chain (16) methodology. To further explore and verify the relationship between phylogenetic and morphological data, outline analysis (a geometric morphometric technique) using elliptic Fourier transforms (17) was also performed on third lower molars (M3) of museum specimens from ISEA and Holocene Sus remains from the site of Liang Bua, Flores (18) [see supporting information (SI) Text].
Identifying the Geographic Origin, Distribution, and Temporal Context of the Pacific Clade
The overall structure of the phylogenetic tree reveals a polytomy of clades and individual haplotypes within a basal cluster (colored gray in Fig. 1), consisting of samples from the Malay Peninsula, Sumatra, Java, Borneo, and the Philippines, suggesting that the origin of the genus Sus occurred within this region (15). This cluster includes all Sus species endemic to ISEA (S. scrofa, S. verrucosus, S. barbatus, S. celebensis, and S. philippensis). A strongly supported branch (posterior probability of 100) leads to a more derived cluster (colored green in Fig. 1) consisting of S. scrofa specimens sampled from mainland Asia and a further strongly supported (posterior probability of 100) “Pacific” clade (colored yellow in Fig. 1). The structure of this tree clearly indicates that the more derived cluster, and thus, the Pacific Clade, must have evolved within mainland Asia. This conclusion is supported by the maximum a posteriori tree (lnL = −4866.94), which places the Pacific Clade in a derived position within the mainland Asian cluster (Fig. 1, green). Because S. scrofa are first identified in the paleontological record during the Early Middle Pleistocene (800–900 kya) at Atapuerca, Spain (19) and Zhoukoudian, China (20), it follows that the original, natural dispersal of Sus out of ISEA into mainland Eurasia (11), and the differentiation of S. scrofa on mainland East Asia, occurred many millennia before pigs were domesticated by humans.
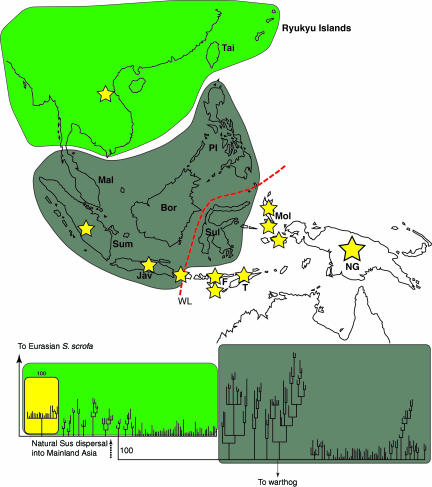
Fig. 1.
Bayesian (Monte Carlo–Markov chain) consensus tree of 249 Sus mtDNA control region haplotypes rooted by a common warthog (Phacochoerus aethiopicus). Gray (basal) and green (derived) clusters on the tree correspond to general regions on the map where the majority of pigs possess haplotypes within that cluster. The yellow clade represents the Pacific Clade. Yellow stars represent islands on which at least one specimen possessing Pacific Clade haplotypes was identified. The yellow star in Southeast Asia represents the two mainland Asian wild boar samples in Vietnam that possessed this haplotype. Regions on the map are: Mal, peninsular Malaysia; Bor, Borneo; Sum, Sumatra; Jav, Java; Sul, Sulawesi; F, Flores; T, Timor (both in the Lesser Sundas); Mol, Moluccas Islands; PI, Philippines; Tai, Taiwan; NG, New Guinea. The dashed red line labeled WL represents the Wallace Line. The black dashed arrow leading to the more derived portion of the tree illustrates the natural movement of wild Sus out of ISEA and into mainland Asia. See SI Fig. 4 for a more detailed version of the tree.
Although numerous mainland modern wild and domestic pigs were sampled from multiple East Asian countries (including Korea, Thailand, Burma, China, Vietnam, and Laos), only two specimens (identified as wild boar on the basis of biometrical evidence) from Vietnam (21) possessed the Pacific signature. These two individuals suggests that the Pacific Clade may have evolved in peninsular Southeast Asia, although additional sampling of the region will be necessary to locate a potential origin.
Although the phylogenetic position of the Pacific Clade confirms its taxonomic distinction and mainland Asian origin, the majority of pigs that form this clade appear to be geographically scattered throughout ISEA and the Pacific. In addition to the two Vietnamese specimens, four Pacific Clade pigs were identified in Sumatra and Java, islands on which indigenous wild populations of Sus existed well before the Neolithic. Pacific Clade pigs also make up 15 of 19 specimens from eight islands east of the Wallace Line in the Moluccas and Lesser Sunda chains and all 17 samples from New Guinea. With the exception of Sulawesi, none of the islands in Wallacea possessed endemic populations of S. scrofa (18, 22–24). In fact, archeological investigations on Flores (18), Timor (24), and the northern Mollucas (13) have demonstrated that the first appearance of pigs is associated with the arrival of the “Neolithic cultural package” during the middle to late Holocene (7000–3500 BP).
The endemic or introduced status and antiquity of the so-called “wild” pigs of New Guinea has been the subject of more debate (8, 25, 26). Our mtDNA data, however, clearly show the ancestry of New Guinea pigs to be directly linked with the dispersal of Pacific Clade pigs, the modern-day (so-called wild) populations most likely being the feral progeny of domesticated individuals originally introduced by farmers to islands east of the Wallace Line.
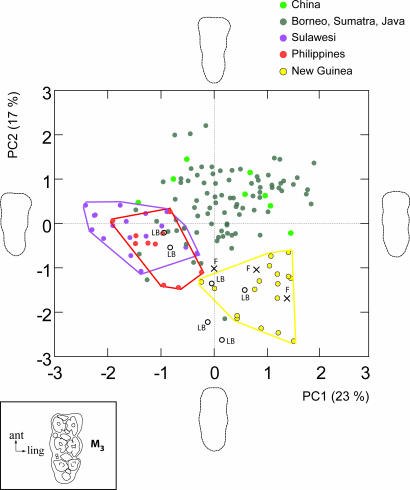
Independent verification of the distinctiveness of pigs with the Pacific mtDNA signature is shown by morphometric analysis of the lower third molar (M3) from the recent New Guinea and Flores pigs and archeological pigs from the site of Liang Bua (Flores) used in our mtDNA studies (see Fig. 3). These data clearly show that pigs from these islands posses a distinct dental morphotype, very different to all other endemic wild ISEA and mainland Sus specimens studied. This morphotype is likely to be correlated with their domestic origins, although further analyses of modern and ancient domestic pig teeth are necessary to confirm this association.
Fig. 3.
Scatter plot of the first two factorial axes of principal components analysis calculated from elliptic Fourier coefficients extracted from third lower molar (M3) occlusal outlines of modern and archaeological pigs from Southeast Asia. The distribution of specimens shows a unique signature for the introduced domestic pigs. The reconstructed outlines depicted on the borders of the plot represent the shape changes corresponding to the component axes. Samples labeled with an F and a cross are modern Sus from Flores, and those labeled with LB and an open circle are archeological pigs from Liang Bua cave also on Flores. The drawing depicts a left third lower molar (M3) in occlusal view (ant, anterior; ling, lingual).
Perhaps more important for assessing the trajectories of human-mediated pig dispersal from mainland East Asia into and within ISEA is the fact that the Pacific signature was absent from samples from Taiwan (which included native wild and domestic modern pigs and an ancient domestic sample), and none of the 40 wild samples from the Philippines (identified as endemic S. philippensis) or the 17 introduced domestic samples from two central Philippine islands, Panay and Cebu. Instead, wild boar from the Philippines form a distinct clade within the basal portion of the tree (see SI Text) alongside western, indigenous ISEA pigs, supporting previous morphological data (27, 28).
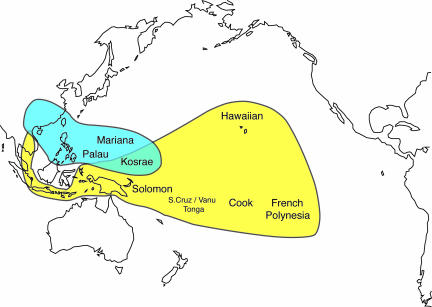
Ancient DNA was successfully extracted from five archeological pig specimens from purportedly pre-European contact sites in the Pacific Islands (from Tubuai, Hanamiai in the Marquesas, and the Tangatatau rock shelter in Mangaia), the Reef Islands (site RF-3), and Mussau (site EKQ), all of which possessed Pacific Clade haplotypes (see SI Text). These ancient sequences therefore unequivocally link Pacific Clade pigs with the Polynesian dispersal (Fig. 2) and by association with that of the earlier Lapita cultural complex, which is associated with the peopling of Remote Oceania (3).
Fig. 2.
Map of East Asia and the Pacific depicting only the distributions of Pacific Clade (yellow) and East Asian haplotypes (light blue). Ancient pigs on the island of Kosrae possess both haplotypes, as do modern pigs from Hawaii, although the East Asian haplotypes on Hawaii are likely the result of postcontact introductions.
Additional Dispersal Episodes Involving Pigs
The mtDNA and morphometric data reveal two additional pig dispersals within Wallacea and into the Pacific that do not involve Pacific Clade haplotypes. The first links East Asia, the Philippines, and Micronesia. The most common Asian haplotypes from mainland East Asia (11) (Fig. 1) are also found in pigs from Taiwan, the Philippines, and the Micronesian islands of Guam and Rota. These haplotypes were also identified in archeological pig specimens from poorly dated contexts in Palau (Fig. 2), probable prehistoric pigs from Kosrae, and an 800- to 1,300-year-old specimen from Taiwan.
Genetic and morphometric analyses of archeological and modern samples from Sulawesi, Flores, and Timor suggest a second dispersal involving an earlier intra-ISEA movement of the Sulawesi warty pig (S. celebensis). A single modern specimen and seven archeological Liang Bua Cave (Flores) specimens (the earliest of which dates to 7000 B.P. based on stratigraphic association and associated 14C dates of charcoal), all possessed a unique haplotype that clusters with modern Sulawesi S. celebensis samples (see SI Text). Assuming that the Holocene distribution of S. celebensis did not naturally extend beyond Sulawesi (18, 22–24), the long-term presence of S. celebensis haplotypes on Flores suggests an early translocation of this species by humans and represents another example of human-mediated animal movement within ISEA and Island Melanesia (25, 29–31). Interestingly, these results also indicate that ancient DNA survival at Liang Bua (site of Homo floresiensis) is possible over long time frames.
Timor is the only other island with pigs in both the Pacific Clade and a S. celebensis clade (albeit a separate lineage from that present on Flores; see SI Text). The data from both islands are consistent with previously reported observations (23) of different species of Sus on these two islands, and with the conclusion that S. celebensis may have been deliberately introduced to Flores in a wild or perhaps even domesticated form (23). These conclusions are also supported by the identification of a distinctive dental morphotype within the range of Eastern ISEA endemic Sus (most likely S. celebensis as also indicated by mtDNA sequences of modern and ancient specimens from Flores) by using morphometric analysis of recent museum and archeological (Liang Bua cave) specimens from Flores (Fig. 3).
Discussion
Our analysis of recent and archeological pig mtDNA and morphometric data clearly suggest that all so-called wild pigs currently found in the lesser Sunda chain and New Guinea (East of the Wallace Line) are descendants of introduced domesticated S. scrofa, which, in turn, trace their mitochondrial genetic heritage to mainland Southeast Asia. Because Pacific Clade haplotypes were found in almost all archaeological pigs we sampled from prehistoric and historic sites in Melanesia and Polynesia, it is clear that Pacific Clade pigs are linked with the main episodes of human dispersal into Near and Remote Oceania.
However, the distribution of Pacific Clade and other Sus haplotypes within mainland Asia, ISEA, and Oceania do not readily support the existing models of Austronesian dispersal. Three modern, fully domestic pigs from Sarawak (Borneo) cluster within the basal portion of the tree (consisting of wild boar from Sumatra, Borneo, and Java). These specimens suggest either that independent pig domestication of Sus sp. occurred in ISEA, or that native, female, wild Sus from Borneo have been crossed recently with pigs domesticated and introduced from East Asia. In either case, because no signatures of basal domestic or wild pigs have yet been identified in ancient or modern pigs from either Wallacea or Oceania, pigs endemic to ISEA likely played no part in Lapita or Polynesian dispersal (Fig. 1).
How can the distribution and frequency of Pacific Clade haplotypes be interpreted? The domestication process itself necessarily selects a small portion of the diversity present in wild populations. Haplotypes associated with that selection are then protected, multiplied, and dispersed by humans. During livestock movement, some domestic pigs are likely to escape and become feral. Because our modern samples derived primarily from wild-caught specimens from museum collections, a low frequency of domestic haplotypes (present in feral pigs) would be expected in regions already possessing endemic suids. However, on islands that have never possessed indigenous Sus populations, so-called wild or native pigs will derive from introduced domestic pigs, subsequently gone feral. This exact pattern is mirrored by the frequency of Pacific Clade pigs, which are found in a small proportion of the total samples from Java and Sumatra, but make up the vast majority of samples from the Lesser Sundas and 100% of pigs sampled from New Guinea.
The complete absence of Pacific Clade haplotypes from modern and ancient specimens from mainland China, Taiwan, the Philippines, Borneo, and Sulawesi suggests that any human dispersal from Taiwan to the New Guinea region via the Philippines, as purported by the “Out of Taiwan” model, did not include the movement of domestic pigs. The origin and trajectory of the pigs associated with both the Lapita cultural complex and the pigs initially taken to Polynesia as part of it must reside elsewhere.
In this context, the initial appearance of pigs in the northern Moluccas after 3500 B.P. (13) is significant. The Neolithic settlers who arrived on these islands and subsequently moved into Oceania must have acquired pigs before this date from somewhere other than Taiwan and the Philippines, most likely in southern Wallacea, a region where significant cultural changes appear to take place during the initial spread of the Neolithic (32), and where our data show high frequencies of introduced domestic pigs exclusively possessing the Pacific signature.
The restricted distribution of Pacific Clade pigs in mainland Southeast Asia, Sumatra, Java, the lesser Sunda islands, and New Guinea poses an interesting additional model for domestic pig (and by proxy human) dispersal, which links the Southeast Asian mainland to the Greater and Lesser Sunda islands and New Guinea. Although not conclusive, the most parsimonious explanation is a west-to-east dispersal trajectory, as supported by the relative frequencies of Pacific Clade haplotypes identified across this distribution.
The phylogenetically based peninsular Southeast Asian origin of the Pacific Clade is further supported by the significant genetic variation present in wild and domestic pigs from northern Vietnam, a pattern previously used to propose an independent center of pig domestication somewhere in peninsular Southeast Asia (21). A recent genetic study of modern chickens (33) also identified Southeast Asia (specifically Vietnam, Burma, and Thailand) as a likely geographic center of early chicken domestication. This finding raises the possibility that the earliest domestic chickens and pigs to arrive in ISEA and Oceania derive from the same geographic source and may have formed part of the same Neolithic dispersal complex.
Conclusions
The data presented here support the existence of two separate, human-mediated dispersals of Sus from Asia into the Pacific and a third within Wallacea. Pigs representing the Pacific Clade originated in East Asia, potentially in peninsular Southeast Asia, where we suggest they were initially domesticated. They were subsequently introduced to the Sunda Islands, the Moluccas, and the New Guinea region. In addition, the Lapita and later Polynesian dispersals into Oceania appear to be exclusively associated with Pacific Clade pigs.
Our findings also highlight the complexities associated with Holocene human migration and the translocation of animal species in ISEA and the Pacific. More complex models allow for various degrees of exchange of language, genes, and artefacts as populations/cultures move from mainland East Asia, through ISEA, Wallacea and Near Oceania and out into the remote Pacific (8, 9, 34). The different components of the Neolithic cultural complex may therefore have different origins and trajectories to Near Oceania where they finally came together and are identified archeologically as Lapita.
Materials and Methods
Modern, Museum, and Ancient Samples.
Of 254 modern and museum specimen individuals from which DNA was extracted, sequences were determined from 243 (SI Table 1) and combined with 512 GenBank entries (SI Table 2) to generate a data set composed of 755 individuals.
An additional 57 bones and teeth from ancient pigs from 27 sites were subjected to DNA extraction techniques, although of those only 23 yielded amplifiable DNA (SI Table 3). A list of haplotype codes and the samples that are represented by each haplotype are found in SI Table 4.
Extraction, DNA Amplification, and Sequencing.
The modern and museum individuals were analyzed in four separate facilities where, in total, 663 bp of mitochondrial control region DNA were amplified and sequenced, although not all of the samples yielded the entire fragment. A total of 145 samples were successfully extracted and amplified at the Henry Wellcome Ancient Biomolecules Centre (HWABC) in Oxford, U.K. Because the DNA preserved within the museum specimens (often >100 years old) was often significantly degraded, all museum specimens were treated as fully ancient material, thus the extraction, amplification, and sequencing protocol at the HWABC followed the ancient DNA methods described by Shapiro et al. (15).
Seventeen modern domestic samples representing six different breeds were extracted at the Institute of Biotechnology, Ha Noi, Vietnam. A total of 73 modern samples were extracted, amplified, and sequenced in the Istituto Nazionale per la Fauna Selvatica in Italy, and 7 modern samples derived from Korean wild boar were extracted at the Animal Genomics and Bioinformatics Division of the National Livestock Research Institute in South Korea. The details of methods used for these samples are discussed SI Text.
A variety of primer combinations were used (SI Table 5) depending on the nature of the sample, and stringent ancient DNA protocols (14), including the use of multiple extraction and PCR blanks, were followed in each laboratory where DNA was extracted from nonmodern samples. In addition, all nonmodern samples were amplified at least two times independently. In total, 41 samples were externally replicated. Cloning reactions were performed at the HWABC using a Topo-TA cloning kit (Invitrogen, Paisley, U.K.) according to the manufacturer's instructions and amplified by using the primers T7 and M13R (Invitrogen). Eight sequences from each cloning reaction were sequenced to evaluate template damage and check for the presence of contaminating sequences and/or numts.
All ancient individuals were analyzed in at least two physically isolated facilities. Given the age and preservation of the ancient samples, only 20 of 47 samples were successfully amplified at the HWABC following the same protocols referenced above (see SI Text).
At the HWABC, four primer combinations (see SI Table 6) were designed to amplify three separate ≈120-bp fragments, each of which contain single nucleotide polymorphisms associated either with specific, geographically linked clusters of haplotypes or, in some cases, with individual haplotypes. Different combinations of all four fragments were amplified in the ancient samples, and every PCR (successful or not) was always independently replicated at least once. In cases where only one or two fragments were successfully amplified for a single sample, phylogenetic analyses (usually neighbor-joining trees) were carried out to identify the subset of haplotypes within the entire range of haplotypes found within the modern and museum samples that matched the ancient sequence. The results of each PCR and the haplotype associations of each successful sample are listed in SI Table 3.
Analysis of eight ancient samples at the Department of Anthropology DNA Laboratories at the University of Auckland followed protocols discussed in detail in SI Text and includes information regarding PCR conditions, primers (SI Table 7), sequencing, replication, and phylogenetic analysis.
Analysis of Sequence Data.
A total of 512 sequences from previous published studies deposited in GenBank were aligned by eye with the 243 new sequences using Se-Al (http://evolve.zoo.ox.ac.uk). Phylogenetic analysis was performed with MrBayes 3 (16), and model parameters were identified by ModelTest (36) (HKY85+G+I). Under this model, parameter estimates (including posterior probabilities) and consensus trees resulting from eight MrBayes runs of at least 10 million (but up to 30 million) generations each were recorded and contrasted. The posterior probabilities listed on the trees represent the lowest recorded values among all of the runs.
Geometric Morphometric Recording and Analyses.
Elliptic Fourier analysis of mandibular third lower molar (M3) outlines was performed on a total of 134 museum Sus specimens from the Natural History Museum (Smithsonian Institution) and the Natural History Museum (Naturalis), Leiden. Three recent specimens from Flores housed in the Indonesian Centre for Archaeology, Jakarta, Indonesia, and six additional ancient specimens from the archeological site of Liang Bua cave (Flores, Indonesia) were also analyzed (SI Table 8).
The rationale for selecting individual teeth and specifically the M3 is listed in SI Text. 2D images of the occlusal outlines were captured with a Coolpix 4500 digital camera (Nikon, Tokyo, Japan). The outline of the molars corresponds to their 2D projection viewed from the occlusal surface.
A total of 100 equally spaced points on each individual outline were semiautomatically sampled and their coordinates recorded using an optical image analyzer (Optimas v.6.2, Optimas Corporation, Bothell, WA). The starting point of the outline was defined at the intersection between the distolingual cusp (entoconid) and the Talonid (hexaconid).
An elliptic Fourier transform was then performed on these coordinates by using NTSYSpc 2.11 (Exter Software, Stauket, NY). This method is based on a separate Fourier decomposition of the incremental changes of the x and y coordinates as a function of the cumulative length along the outline. Each function (x and y variations) was decomposed into a sum of trigonometric functions of decreasing wavelength (i.e., harmonics). Hence, each harmonic corresponds to four coefficients: An and Bn for x and Cn and Dn for y, defining an ellipse in the xy plane. The coefficients of the first harmonic, describing the best-fitting ellipse to the original outline, were used to standardize the size, orientation, and starting point of the molar outlines. These coefficients correspond to residuals after standardization and should not be included in following statistical analyses (37).
Principal component analysis was performed on 116 coefficients (harmonics 2–30) by using NTSYSpc 2.11. Visualization of molar shape change along the principal component axes was performed by using multivariate regression as suggested by Rohlf and Archie (35). Shape changes are depicted by reconstructed outlines. An outline can be reconstructed from any set of Fourier coefficients following the inverse Fourier method using NTSYSpc 2.1.1.
Supplementary Material
Acknowledgments
We thank the many institutions and individuals that provided sample material and access to collections and Kristofer Helgen for comments on the manuscript. This work was supported by the Wellcome Trust (K.D.), the Leverhulme Trust (G.L.), the Smithsonian Institution Museum of Natural History (K.D.), and the Fyssen Foundation (T.C.).
Abbreviations
- ISEA
Island Southeast Asia
- HWABC
Henry Wellcome Ancient Biomolecules Centre.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ779287–DQ779542, DQ779544–DQ779551, and DQ841948–DQ841949).
This article contains supporting information online at www.pnas.org/cgi/content/full/0607753104/DC1.
References
- 1.Diamond JM. Nature. 2000;403:709–710. doi: 10.1038/35001685. [DOI] [PubMed] [Google Scholar]
- 2.Bellwood P. In: Archaeology and Language II: Archaeological Data and Linguistic Hypotheses. Blench R, Spriggs M, editors. London: Routledge; 1998. pp. 128–140. [Google Scholar]
- 3.Kirch PV. On the Road of the Winds: An Archaeological History of the Pacific Islands Before European Contact. Berkeley, CA: Univ California Press; 2000. [Google Scholar]
- 4.Bellwood P, Diamond J. World Archaeol. 2005;37:503–506. [Google Scholar]
- 5.Terrell JE, Kelly KM, Rainbird P. Curr Anthropol. 2001;42:97–124. [Google Scholar]
- 6.Oppenheimer S. World Archaeol. 2004;36:591–600. [Google Scholar]
- 7.Hurles ME, Matisoo-Smith E, Gray RD, Penny D. Trends Ecol Evol. 2003;18:531–540. [Google Scholar]
- 8.Green RC. In: Australian Archaeologist: Collected Papers in Honor of Jim Allen. Anderson A, Murray T, editors. Canberra, Australia: Coombs Academic; 2000. pp. 372–392. [Google Scholar]
- 9.Anderson AJ. J Austronesian Stud. 2005;1:27–48. [Google Scholar]
- 10.Matisoo-Smith E, Robins JH. Proc Natl Acad Sci USA. 2004;101:9167–9172. doi: 10.1073/pnas.0403120101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larson G, Dobney K, Albarella U, Fang MY, Matisoo-Smith E, Robins J, Lowden S, Finlayson H, Brand T, Willerslev E, et al. Science. 2005;307:1618–1621. doi: 10.1126/science.1106927. [DOI] [PubMed] [Google Scholar]
- 12.Allen MS, Matisoo-Smith E, Horsburgh A. Int J Osteoarchaeol. 2001;11:4–13. [Google Scholar]
- 13.Bellwood P, White P. Science. 2005;309:381. doi: 10.1126/science.309.5733.381a. and author reply (2005) 309:381. [DOI] [PubMed] [Google Scholar]
- 14.Cooper A, Poinar HN. Science. 2000;289:1139. doi: 10.1126/science.289.5482.1139b. [DOI] [PubMed] [Google Scholar]
- 15.Shapiro B, Drummond AJ, Rambaut A, Wilson MC, Matheus PE, Sher AV, Pybus OG, Gilbert MTP, Barnes I, Binladen J, et al. Science. 2004;306:1561–1565. doi: 10.1126/science.1101074. [DOI] [PubMed] [Google Scholar]
- 16.Ronquist F, Huelsenbeck JP. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 17.Renaud S, Michaux J, Jaeger JJ, Auffray JC. Paleobiology. 1996;22:255–265. [Google Scholar]
- 18.Morwood MJ, Soejono RP, Roberts RG, Sutikna T, Turney CSM, Westaway KE, Rink WJ, Zhao JX, van den Bergh GD, Due RA, et al. Nature. 2004;431:1087–1091. doi: 10.1038/nature02956. [DOI] [PubMed] [Google Scholar]
- 19.van der Made J. J Hum Evol. 1999;37:389–413. doi: 10.1006/jhev.1998.0264. [DOI] [PubMed] [Google Scholar]
- 20.Kurtén B. Pleistocene Mammals of Europe. London: Weidenfeld & Nicolson; 1968. [Google Scholar]
- 21.Hongo H, Ishiguro N, Watanobe T, Shigehara N, Anezaki T, Long VT, Binh DV, Tien NT, Nam NH. Zool Sci. 2002;19:1329–1335. doi: 10.2108/zsj.19.1329. [DOI] [PubMed] [Google Scholar]
- 22.Groves CP. Ancestors for the Pigs: Taxonomy and Phylogeny of the Genus Sus. Canberra, Australia: Australian Natl Univ; 1981. technical bulletin 3. [Google Scholar]
- 23.Groves CP. J Soc Oceanistes. 1983;77:105–119. [Google Scholar]
- 24.Glover I. Archaeology in Eastern Timor, 1966-67. Canberra, Australia: Terra Australis II; 1986. [Google Scholar]
- 25.Spriggs MJT. The Island Melanesians. Cambridge, MA: Blackwell; 1997. [Google Scholar]
- 26.Allen J. Mod Q Res Southeast Asia. 2000;16:137–176. [Google Scholar]
- 27.Lucchini V, Meijaard E, Diong CH, Groves CP, Randi E. J Zool. 2005;266:25–35. [Google Scholar]
- 28.Groves CP. Zool J Linnean Soc. 1997;120:163–191. [Google Scholar]
- 29.Flannery T. Mammals of the South-West Pacific and Moluccan Islands. Ithaca, NY: Cornell Univ Press; 1995. [Google Scholar]
- 30.Flannery T, Kirch PV, Specht J, Spriggs M. Archaeol Oceania. 1988;23:89–94. [Google Scholar]
- 31.Flannery TF, White JP. Res Explor. 1991;7:96–113. [Google Scholar]
- 32.Spriggs M. Rev Archaeol. 2003;24:57–80. [Google Scholar]
- 33.Liu YP, Wu GS, Yao YG, Miao YW, Luikart G, Baig M, Beja-Pereira A, Ding ZL, Palanichamy MG, Zhang YP. Mol Phylogenet Evol. 2006;38:12–19. doi: 10.1016/j.ympev.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 34.Green RC. In: Indo Pacific Prehistory. Bellwood P, editor. Vol 2. Canberra, Australia: Indo-Pacific Prehistory Assoc; 1991. pp. 295–305. [Google Scholar]
- 35.Rohlf FJ, Archie JW. Syst Zool. 1984;33:302–317. [Google Scholar]
- 36.Posada D, Crandall KA. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 37.Crampton JS. Lethaia. 1995;28:179–186. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.