Abstract
What determines which plant species are susceptible to a given plant pathogen is poorly understood. Experimental inoculations with fungal pathogens of plant leaves in a tropical rain forest show that most fungal pathogens are polyphagous but that most plant species in a local community are resistant to any given pathogen. The likelihood that a pathogen can infect two plant species decreases continuously with phylogenetic distance between the plants, even to ancient evolutionary distances. This phylogenetic signal in host range allows us to predict the likely host range of plant pathogens in a local community, providing an important tool for plant ecology, design of agronomic systems, quarantine regulations in international trade, and risk analysis of biological control agents. In particular, the results suggest that the rate of spread and ecological impacts of a disease through a natural plant community will depend strongly on the phylogenetic structure of the community itself and that current regulatory approaches strongly underestimate the local risks of global movement of plant pathogens or their hosts.
Keywords: fungal pathogen, plant disease ecology, tropical forest, plant quarantine, host specificity
Most species of plant pathogen can attack a broad diversity of plant species (1), but the number of plant species with which a pathogen interacts in a local community is generally much lower (2). However, we have limited abilities to predict which species within a plant assemblage are most likely to be susceptible to a particular pathogen. Existing databases of pathogen–host range (e.g., ref. 1) have limited value for quantitative assessments because they are based primarily on haphazard records of pathogens on economically important plants. More importantly, only plant species that are susceptible to particular pathogens were recorded but not which plants are resistant. Nevertheless, several economically and scientifically important issues require predicting the likely host range of plant pathogens. The idea of selectivity among local plant species underlies the theory for the role of natural enemies in the maintenance of plant diversity (3–5) and biological invasions (6, 7). Host selectivity is used in studying plant disease epidemics (8, 9), estimating fungal biodiversity (10), managing agriculture and forestry systems (11, 12), and in risk analysis for global movement of plants and pathogens (13, 14).
Conventional wisdom is that two closely related plant species should be more likely to be susceptible to the same plant pathogens than would plants that are evolutionarily distant, because the morphological and chemical traits of plants that regulate interactions with pathogens are often phylogenetically conserved (15). Indeed, recent work has shown a strong phylogenetic signal in host range of herbivorous insects in tropical rain forest (16, 17), and there is qualitative support for such a signal for fungal pathogens (7). The presumption of phylogenetic signal in host range often underlies important agronomic and economic decisions. For instance, risk assessment for the release of exotic natural enemies for biological control of weedy plants is predicated on host specificity. Empirical host-range testing follows the “centrifugal phylogenetic method” (14, 18–20), with testing being most intense on local species in the same genus as the target host, less intense in the same family, and with still fewer test on hosts at greater phylogenetic distances. In a second example, the U.S. regulatory Animal and Plant Health Inspection Service (APHIS) has developed a draft policy whereby exotic plant species from the same genus as a known host of a quarantine pathogen would not be authorized for planting in the U.S. pending further risk analysis (13). The APHIS policy uses a “step–function” phylogenetic model of pathogen host range, where plants outside the genus are not considered to pose a risk for pathogen spread. However, neither of these applications of phylogenetic signal is based on robust empirical data. Evaluating and improving these models requires quantitative estimates of the strength and shape of phylogenetic signal in the host range in plant pathogens (21, 22), based on empirical evaluation of both susceptibility and resistance of phylogenetically diverse plant species to a diversity of plant pathogens.
We used experimental, in situ inoculations to evaluate the host range of 53 necrotrophic (tissue-killing) plant pathogenic fungi on a diversity of plant species in two experiments in Panama: first, in an artificial assemblage of tree species in a reforestation nursery (Nursery), and second in a natural assemblage of species in a semideciduous lowland moist tropical forest (Forest). The Forest inoculations permit assessment under conditions where historical plant–pathogen interactions may have led to local natural selection on either hosts or pathogens (or both) at a particular site, whereas the Nursery inoculations permit phylogenetic analysis separate from such local rapid evolutionary responses. We analyzed the likelihood that a pathogen would cause disease on a target host as a function of the phylogenetic distance between the target plant and the plant species from which the pathogen was isolated. This is a systematic test of the phylogenetic signal in the host range of foliar plant pathogens.
Results
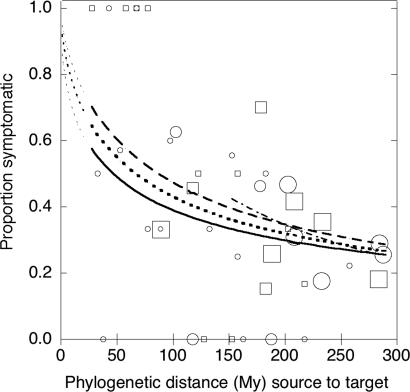
In both Nursery and Forest studies, the proportion of plant species that developed disease declined continuously with phylogenetic distance (estimated time of independent evolution) between the source and target host species from 28 to 287 My (Fig. 1), with the steepest decline occurring in the most closely related pairs. The responses were similar in the two studies (slopes not different, P = 0.6), whether the test was performed with a haphazard collection of plant species (Nursery), or with a natural plant community (Forest), where reciprocal selection for pathogenicity and susceptibility would likely have occurred between pathogens and plants over many generations. In our taxa, congeneric pairs (n = 9) were separated by a median distance of 53.6 My (range: 28–54 My), with 66.7% of the cross-inoculations causing disease, and confamilials (n = 94) were separated by a median distance of 85 My (range: 34–135), with disease in 43.6% of cross-inoculations, whereas the remaining, more distant pairings (n = 859) were separated by a median distance of 233 My (range: 50–288), with 29.9% susceptible. Even when all plant pairs with phylogenetic distances <135 My were removed from analysis (including all congeneric and confamilial pairs), there was still a highly significant effect of phylogenetic distance on the likelihood of symptom development (Fig. 1). Thus, the phylogenetic signal in pathogen–host range is not lost at phylogenetic distance beyond taxonomic levels like genus and family but extends to ancient phylogenetic distances between unrelated angiosperms.
Fig. 1.
Proportion of target plant species that developed disease symptoms after inoculation with fungal pathogens from source plant species. Logistic regressions were performed on results of individual pathogen–host combinations, but data were grouped into 5-My source-target phylogenetic distance classes for illustration. Circles (Forest study) and squares (Nursery study) indicate the proportion of inoculations within a 5-My class that produced disease symptoms. Lines indicate the predicted proportion symptomatic based on logistic regression for the Forest study (solid line), the Nursery study (dashed line), or all data combined (dotted line). Fine dotted lines show extrapolation of predictions to shorter phylogenetic distances than represented by experimental data. Size of symbol indicates the number of inoculation pairs included in the distance class (grouped as <10, 10–30, or >30 inoculations). Logistic regressions were performed on raw (not binned) data. The logistic fit still shows slope when pairs of <135 My are excluded (dash–dot line). The logistic regression formulas are: forest: logit(S) = 2.2327 − 1.3428 × (log10(distance + 1)), χ2 = 6.96, P = 0.0084, n = 578; Nursery: logit(S) = 3.4096 − 1.7562 × (log10(distance + 1)), χ2 = 7.66, P = 0.0056, n = 384; Combined: logit(S) = 2.9113 − 1.5944 × (log10(distance + 1)), χ2 = 16.71, P = 0.0001, n = 962; and combined, >135 My: logit(S) = 5.93699 − 2.85376 × log10(distance + 1); χ2 = 8.09, P = 0.004, n = 821; where Prob(symptomatic) = exp(logit(S))/[1 + exp(logit(S)].
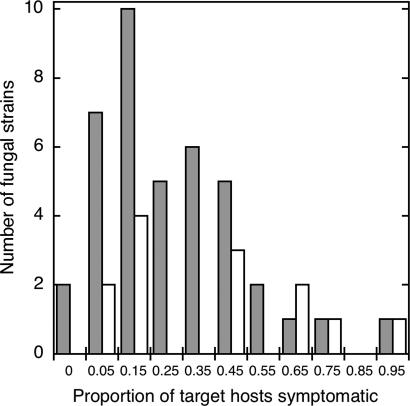
We currently lack good general estimates for breadth of host range for necrotrophic plant pathogenic fungi. Our results suggest that most pathogens in a tropical forest are pathogenic on a large number of locally available hosts but that most local plant species will be resistant. Only 2 of the 53 tested pathogens (3.8%) were restricted to a single host species (Fig. 2). Most species had a moderate number of potential host species (median 27.7% of host species tested), with a few pathogens appearing to be very broad generalists (Fig. 2).
Fig. 2.
Frequency distribution of observed host breadth. Shown is the proportion of target hosts that produced symptoms after inoculation, excluding the original source host species, for 40 fungal pathogens in the forest (gray bars), and 13 in the Nursery (white bars). Each strain was tested on 11–36 host species (median 16). Only two strains (of 53) appeared to be specific to one plant species (n = 11 and 15 hosts tested). For all strains combined, mean host range breadth = 0.311 ± 0.241, median = 0.2727.
Discussion
The phylogenetic signal we measured in pathogen–host range has significant applications in numerous areas; here, we outline the implications for epidemiology, ecology, biodiversity, agronomy, and quarantine risk analysis.
The rate of spread of a fungal plant pathogen increases with the density of suitable hosts (23); for pathogens able to infect multiple species, this density is, in turn, a function of the number of susceptible species in a community. Each plant species in the forest will be linked to others through polyphagous pathogens, and the number of “cohosts” in a local forest will be a function of the phylogenetic structure of plant species in the community. We estimated how many host species in a particular forest would be expected to be susceptible to a hypothetical pathogen with a particular host, using the phylogenetic structure of the 315 species in the well studied, nearby 50-hectare (ha) Forest Dynamics Plot on Barro Colorado Island and the phylogenetic signal estimated in our Forest study (Fig. 1). For a host species with a large number of close relatives on the plot (e.g., Inga marginata, Fabaceae, 25% quartile phylogenetic distance to all other heterospecifics = 188.5 My), there were 14 other species (4.5% of pool) with which it had a >50% chance of having a pathogen in common. In contrast, for a species with few close relatives (e.g., Attalea butyraceae, Arecaceae, 25% quartile distance = 287.7 My), there were no other species on the plot with such a high likelihood. At the 30% probability level for having a pathogen in common, there were 115 likely cohosts for pathogens of Inga compared with only 10 for Attalea. The variation in likely number of cohosts across plant species demonstrates the potential for ecologically significant differences in disease epidemiology depending on the phylogenetic structure of the plant community.
Plant pathogens with limited local host ranges are thought to help maintain plant diversity in forest communities (4, 24). In our study, most local plant species were resistant to any given pathogen, but species-specialists or even genus-specialists were rare. Analyzing tropical herbivore communities, Novotny et al. (17) found that a lack of strict host specificity and gradual decrease in insect species overlap with phylogenetic distance between plant pairs. Most herbivores fed on several closely related congeneric plant species, but many fed on plants from multiple genera and families. Our data suggest that, because close relatives are more likely to be susceptible to the same pathogens, pathogens may be most effective at maintaining diversity of higher taxonomic levels [“phylodiversity”(22)] rather than species diversity per se. An important next step is to evaluate how the severity of damage varies among pathogens of different host ranges.
Efforts to estimate biodiversity have often used extrapolations based on estimates of host specificity of insects or fungi (10, 25). Incorporation of the phylogenetic signal in host sharing in insects (simplified to 100% host sharing in herbivorous insects for all plants of the same genus), led to significantly reduced estimates of global insect diversity compared with earlier efforts (17). Our more sensitive measure of host breadth provides a continuous model for estimating fungal diversity. Such calculations would require strong (and not yet available) estimates of pathogen diversity within individual species but should significantly reduce current estimates of global fungal diversity.
In designing mixed-species forestry plantations or intercropped agronomic systems (11, 26), the mixing of host species that are phylogenetically distant should decrease the probability that hosts would share common pathogens, effectively reducing apparent host density for density-dependent pathogens. In our system, species pairs with phylogenetic distances of >250 My would reduce by half the probability of hosts sharing pathogens compared with mixtures of species from the same family. In an agricultural example, when maize (Zea mays) was intercropped with the distantly related (288 My) common bean (Phaseolus vulgaris), the incidence of common rust (Puccinia sorghi) on maize was greatly reduced, whereas maize suffered high rust incidence when intercropping with the confamilial sorghum (Sorghum bicolor) (55 My) (11). The causes for differences in disease levels in the two systems were not determined in the study, but Sorghum has been reported as a host for the pathogen (1).
Finally, the observed data provide a quantitative assessment of the assumption of a phylogenetic signal in host range used in risk assessment for biological control agents, biological invaders, and quarantine decisions. Our data suggest that arbitrary cutoffs at the genus or family level will underestimate host ranges of plant pathogens and their associated risks. Analysis of likely hosts based on a continuous logistic function of estimated phylogenetic distance may provide a more realistic evaluation of risks from introduced pathogens.
These results represent a quantitative assessment of phylogenetic signal in the host range of plant pathogens. They provide a benchmark for evaluating the robustness of existing tools for understanding the spread, impacts, and evolutionary biology of plant pathogens as well as the basis for development of novel predictive tools in plant ecology and risk analysis.
Methods
Study Sites.
We evaluated the phylogenetic signal of host range of a 53 diverse plant pathogenic fungi in two studies in central Panama. In the first study (Nursery), we used seedlings of 45 species of tropical forest trees grown for reforestation projects at the PRORENA nursery in Gamboa, Colón Province [09°07.225N, 079°42.242W, 73 meters above sea level (masl)]. Although all of the species in the nursery are native to forests in Panama, they are not a natural assemblage, permitting a test of host range in the absence of local selection pressure for particular plant–pathogen pairs. No more than half of the nursery species have been reported in any one of the three large forest plots located in central Panama (midisthmus Barro Colorado Island, 50 ha; Caribbean side Fort Sherman, 6 ha; Pacific side Cocoli, 4 ha) (http://ctfs.si.edu/), and 31% of the species were not reported in any of the three plots. Plants were grown under partial shade with overhead misting.
In the second study (Forest), three plots (50-m radius, 0.785 ha) were located in the forest near the town of Gamboa and along Pipeline Road, in the Parque Nacional Soberanía, Colón Province, Republic of Panama. The forest in this area is semideciduous, lowland tropical moist forest. Here, host range testing was conducted on species from within the immediate neighborhood of pathogen isolation where past selection may have shaped the host ranges of local pathogens. The three Forest plots were all separated from each other by at least 1 km (Plot 1 09°07.258N, 079°41.840W, 80 masl; Plot 2 09°08.213N, 079°43.448W, 75 masl; Plot 3 09°09.026N, 079°43.987W, 89 masl). Annual precipitation in Gamboa is 2,133 mm, with 87% of the rain falling between May and November (Panama Canal Authority data, 1897–2005, http://striweb.si.edu/esp/physical_monitoring/index_phy_mon.htm). All of the forest sites have histories of some anthropogenic disturbance dating to canal construction, but all show a mature, multilayered canopy structure.
Plots were systematically surveyed for plant composition in the understory and overstory. Plant identifications were based on comparison with specimens in the herbaria at the Smithsonian Tropical Research Institute and the University of Panama. Plant taxonomy follows that of Correa et al. (27), except where in conflict with the Angiosperm Phylogeny Group (28). Vouchers of unusual species have been deposited with the herbarium of the University of Panama.
Pathogen Isolation.
We evaluated the phylogenetic signal of host range of a diversity of plant pathogenic fungi in two studies in central Panama. We focused on culturable, nectrotrophic fungal pathogens to facilitate cross-inoculations, and because biotrophic pathogens like rusts and smuts are very rare in tropical forests (29) and were not observed at our sites. In both the Nursery and Forest sites, representative leaves of each disease type were collected, placed in plastic bags, and returned to the laboratory for processing within 1 h. Two triangular pieces of tissue were collected from the edge of the symptomatic area by using a hole punch for mounting small insects (area, 7 mm2). These leaf pieces were placed together in a mesh tea strainer spoon and immersed for 1 min in 70% ethanol and then for 1 min in 10% commercial bleach (0.525% sodium hypochlorite) to surface sterilize. They were then transferred to malt extract agar with chloramphenicol (MEA-chlor: 2% malt extract, 1.5% agar, 0.02% chloramphenicol), and incubated at ambient air-conditioned (≈23°C) laboratory conditions. After 3–4 days, fungal growth from the leaf pieces was transferred to a new plate of MEA. If multiple fungal morphotypes were apparent, each was separately transferred. Pathogens were isolated during the rainy season: in August 2005 at the Nursery, and in November and December 2005 for the Forest pathogens.
In the first study (Nursery), we isolated into pure culture 13 strains of fungi from diseased leaves of 12 species of plants within the Nursery. In the second study (Forest), we isolated 40 foliar fungal pathogens from a diversity of plants in each of three plots (12 pathogens from 11 plant species in Plot 1, 16 from 16 species in Plot 2, and 12 from 12 species in Plot 3). These fungi include at least 19 species from 11 genera in 6 orders of Ascomycetes. A list of the fungi used in these experiments and the hosts from which they were isolated is available electronically in supporting information (SI) Data Set 1. Each fungal strain was preserved by placing mycelial plugs in 2-ml cryovials and covering with sterile water and by growing in slant-culture in glass vials on MEA. Dried fungal cultures as well as pressed, symptomatic leaves of the original source plants were deposited as vouchers in the herbarium of the University of Panama.
Pathogen Inoculum Preparation.
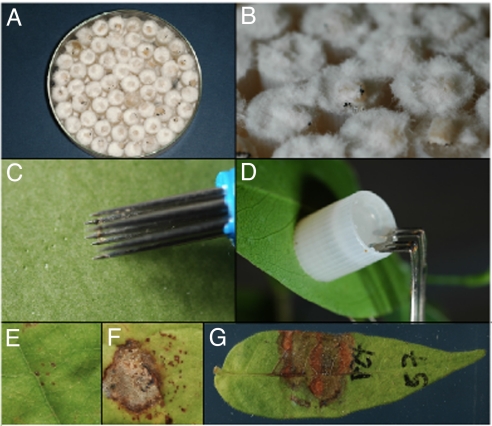
Caps of 2-ml external-thread cryovials (Nalge, Naperville, IL) were placed in either deep, glass Petri dishes or in aluminum baking pans (with aluminum foil cover) with the deep end of the cryovial facing upward. The cryovial caps were autoclaved for 15 min. Into each sterile cryovial cap, we then placed a small piece (≈2–4 mm2) of colony cut from 1-wk-old fungal cultures grown on MEA. Sterile, molten, cooled (≈56°C) MEA was poured carefully into each cap from a sterile graduated cylinder until the cap was completely full. The trays were recovered with the sterile aluminum foil to allow caps to solidify. For initial inoculations in the PRORENA experiment, the fungi were allowed to grow in the caps in the Petri plates (Fig. 3 A and B), but for most inocula, once the agar had solidified, the caps were removed from the trays by using sterile forceps, and placed into sterile Whirl-pak bags (Nasco, Fort Atkinson, WI) and sealed. The cap cultures were incubated in ambient air-conditioned (≈23°C) laboratory conditions for 7–10 days before use for inoculation. Bags were shaken every other day to prevent mycelium from binding the caps together.
Fig. 3.
Inoculation procedure for testing whether fungal pathogens can cause disease on leaves. (A) Pathogen inoculum grown in agar-filled cryovial caps. (B) Closeup of inoculum. (C) Wounding the leaf surface. (D) Clamping the inoculum to the leaf. (E) Wound response (resistant). (F) Necrosis (susceptible). (G) Diseased leaf 7 d after inoculation.
Inoculation Methods.
Leaves selected for inoculation were labeled with permanent marker on the upper surface with the plant number (at the tip of the leaf) and the strain number (e.g., P72). Small wounds were made adjacent to the label (Fig. 3C), by lightly touching the underside of the leaf twice with a seven-pointed Pergamano flower tool (Cat. no. 1111; Pergamano International, Uithoorn, The Netherlands); this results in 14 pin pricks in an area of ≈0.3 cm2. Leaves were wounded because most foliar pathogens in tropical forests require wounds to successfully cause disease (30). The inoculum cap was removed from the Whirl-pak bag by using forceps (soaked in 70% EtOH between uses), pressed against the wound on the underside of the leaf, and clipped in place with a bent hair clip (Goody, Atlanta, GA) (Fig. 3D). Control inoculations were caps filled with sterile MEA. One week after inoculation, leaves were harvested and photographed, and symptoms were recorded. The interactions were recorded as “susceptible” if clear disease symptoms developed after inoculation, but control inoculations showed either only minimal wound reaction (usual) or a very different type of disease symptom (Fig. 3 E–G). When symptoms were similar on control and inoculated leaves, the inoculation was excluded from the data set. Seven pathogens (one from the nursery and six from forest plots) that never caused disease symptoms (even on original host species) were excluded from analysis. Nursery inoculations were performed in the rainy season 2005; Forest inoculations were performed in the early dry season 2006.
For each pathogen, we tested for pathogenicity on a range of species using in situ inoculations onto plants within the plot of origin only, so that (i) in each plot, each pathogen was inoculated onto the source host for all pathogens from that plot (full reciprocal inoculations), and (ii) each pathogen was inoculated onto those plant species that were most closely related to the source plant species (phylogenetic distance <≈170 My). Host selection was therefore intentionally biased to include more close relatives than expected at random in the forest. Each pathogen was inoculated onto 11–17 plant species (mean 14.5). In the Nursery, each pathogen was inoculated onto seedlings of 19–36 target species (mean 29.5, not including original host species). Overall, there were 384 fungus-plant pairs in the Nursery study and 578 pairs in the Forest study. In situ inoculations mean that the inoculated leaves were already colonized by a large diversity of endophytic fungi (31), so that inoculated strains interacted with the host plants in their natural state of plant-fungus chimeras (32).
Analyses.
To estimate phylogenetic distances between plant species, we first created an hypothesis for the phylogenetic relationships among our plant species based on the dated angiosperm supertree of Davies et al. (33), using the desktop version of Phylomatic (34). This tool joins sampled species at the appropriate place to a larger phylogenetic hypothesis, maintaining the branch lengths in the base tree, and prunes all intervening taxa. In the absence of information about intrafamilial phylogenetic resolution, relationships are modeled as polytomies. The resultant tree was ultrametric, with branch lengths reflecting estimated time between branching events. The Phydist function of Phylocom (v. 3.34b) was used to extract pairwise phylogenetic distances (or time of independent evolution, in My) between all plant species. Conspecific inoculations (onto the host species of origin) were excluded from analysis to avoid a biased estimate of host sharing at short phylogenetic distances. Results from the three forest plots were combined for analysis.
For the Nursery and Forest experiments separately (and combined), we fit a logistic regression model using log10(phylogenetic distance + 1) as the independent variable, and the host response (susceptible or resistance) as the dependent nominal variable. Analyses were performed by using JMP (v 5.1.2, SAS Institute, Cary, NC).
To estimate the likelihood that host would be susceptible to the same pathogens in a lowland rain forest, we used the species lists from the well studied 50-ha Forest Dynamics Plot on Barro Colorado Island (refs. 35 and 36 and Condit, R., Hubbell, S. P., and Foster, R. B., Barro Colorado Island Forest Census Plot Data, http://ctfs.si/edu/datasets/bci), some 4 km from our plots. We used Phylomatic and Phylocom (Phydist) with the same Davies et al. (33) supertree to estimate the phylogenetic distance between each pair of trees for the 315 woody plant species recorded from the plot. For each species on the plot, we then calculated the 25% quartile of distances to all other 314 species. We then used the logistic regression equation from the Forest plots (Fig. 1) to determine the probability that a pathogen from each host would also be pathogenic on each of the other 314 tree species and tallied the number of species in the plot with a predicted likelihood >0.3 or >0.5 of being susceptible to a pathogen from the focal host.
Supplementary Material
Acknowledgments
We thank J. Deago, W. Jácome, and M. Wishnie of the Native Species Reforestation Project (PRORENA) for providing seedlings, space, and logistical support; A. Bethancourt, N. Florez, H. Membache, and J. Sarco for assistance in the laboratory and field; R. Aizprúa (Flora Tropical, S.A.) and R. Pérez [Smithsonian Tropical Research Institute (STRI)] for plant identifications and plot mapping; STRI and the Center for Tropical Forest Science for logistical support; B. Ayala, K. Garrett, A. Jani, B. Lyon, I. M. Parker, and Y. Springer for discussions and comments on the manuscript; and the Republic of Panama for conserving their forests and making them available for study. This work was supported by National Science Foundation Grants DEB 0515520 and DEB 0212873, Pacific Rim Foundation Grant 05-1487, and the University of California, Santa Cruz.
Abbreviations
- ha
hectare
- masl
meters above sea level
- MEA
malt extract agar.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607968104/DC1.
References
- 1.Farr DF, Rossman AY, Palm ME, McCray EB. Fungus–Host Distributions, Fungal Databases, Systematic Botany and Mycology Laboratory. 2004 (Agric Res St/US Dep Agric) http://nt.ars-grin.gov/fungaldatabases/
- 2.Thompson JN. The Geographic Mosaic of Coevolution. Chicago: Univ of Chicago Press; 2005. [Google Scholar]
- 3.Gillett JB. Syst Assoc Pub No. 1962;4:37–46. [Google Scholar]
- 4.Janzen DH. Am Nat. 1970;104:501–527. [Google Scholar]
- 5.Connell JH. In: Dynamics of Numbers in Populations (Proceedings of the Advanced Study Institute, Osterbeek 1970. Boer PJ, Graadwell GR, editors. The Netherlands: Cent Agric Publ Doc Wageningen; 1971. pp. 298–312. [Google Scholar]
- 6.Keane RM, Crawley MJ. Trends Ecol Evol. 2002;17:164–170. [Google Scholar]
- 7.Parker IM, Gilbert GS. In: Annual Review of Ecology, Evolution, and Systematics. Futuyma D, Shaffer HB, Simberloff D, editors. Vol. 35. Palo Alto, CA: Annu Rev; 2004. pp. 675–700. [Google Scholar]
- 8.Altizer S, Hervell D, Friedle E. Trends Ecol Evol. 2003;18:589–596. [Google Scholar]
- 9.Garrett KA, Mundt CC. Phytopathology. 1999;89:984–990. doi: 10.1094/PHYTO.1999.89.11.984. [DOI] [PubMed] [Google Scholar]
- 10.Hawksworth DL. Mycol Res. 2001;105:1422–1432. [Google Scholar]
- 11.Fininsa C, Yuen J. Crop Protection. 2001;20:669–678. [Google Scholar]
- 12.Schroth G, Krauss U, Gasparotto L, Aguilar JAD, Vohland K. Agrofor Syst. 2000;50:199–241. [Google Scholar]
- 13.Animal and Plant Health Inspection Service. Host Status of Commodity Import Analysis and Operations (CIAO), Plants for Planting Imports and Policy, Draft of October 2005, Phase 1 of Q37 revision. 2005. (APHIS, US Dep Agric)
- 14.Wapshere AJ. Ann Appl Biol. 1974;77:201–211. [Google Scholar]
- 15.Farrell BD. Molec Phylogen Evol. 2001;18:467–478. doi: 10.1006/mpev.2000.0888. [DOI] [PubMed] [Google Scholar]
- 16.Novotny V, Basset Y. Proc R Soc London Ser B. 2005;272:1083–1090. doi: 10.1098/rspb.2004.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Novotny V, Basset Y, Miller SE, Weiblen GD, Bremer B, Cizek L, Drozd P. Nature. 2002;416:841–844. doi: 10.1038/416841a. [DOI] [PubMed] [Google Scholar]
- 18.Weidemann GJ, Tebeest DO. Weed Technol. 1990;4:465–470. [Google Scholar]
- 19.Briese DT. In: Improving the Selection, Testing and Evaluation of Weed Biological Control Agents. Technical Series 7. Jacob HS, Briese DT, editors. Glen Osmond, Australia: Coop Res Cent Aust Weed Manage; 2003. pp. 23–33. [Google Scholar]
- 20.Barton J. Biol Control. 2004;31:99–122. [Google Scholar]
- 21.Mitchell CE, Agrawal AA, Bever JD, Gilbert GS, Hufbauer RA, Klironomos JN, Maron JL, Morris WF, Parker IM, Power AG, et al. Ecol Lett. 2006;9:726–740. doi: 10.1111/j.1461-0248.2006.00908.x. [DOI] [PubMed] [Google Scholar]
- 22.Webb CO, Gilbert GS, Donoghue MJ. Ecology. 2006;87(Suppl):S123–S131. doi: 10.1890/0012-9658(2006)87[123:psmssa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 23.Burdon JJ, Chilvers GA. Annu Rev Phytopathol. 1982;20:143–166. [Google Scholar]
- 24.Packer A, Clay K. Nature. 2000;404:278–281. doi: 10.1038/35005072. [DOI] [PubMed] [Google Scholar]
- 25.Erwin TL. Coleopt Bull. 1982;36:74–75. [Google Scholar]
- 26.Lygis V, Vasiliauskas R, Stenlid J, Vasiliauskas A. For Ecol Manag. 2004;201:275–285. [Google Scholar]
- 27.Mireya D, Correa A, Caldames C, de Stapf MS. Catalogo de las Plantas Vasculares de Panama. Bogota, Colombia: Quebecor World Bogota; 2004. [Google Scholar]
- 28.Angiosperm Phylogeny Group (APG)2. Bot J Linn Soc. 2003;141:399–436. [Google Scholar]
- 29.García-Guzmán G, Morales E. Ecology. 2007 doi: 10.1890/05-1174. in press. [DOI] [PubMed] [Google Scholar]
- 30.García-Guzmán G, Dirzo R. Am J Bot. 2000;88:634–645. [PubMed] [Google Scholar]
- 31.Arnold AE, Maynard Z, Gilbert GS. Mycol Res. 2001;105:1502–1507. [Google Scholar]
- 32.Herre EA, Van Bael SA, Maynard Z, Robbins N, Bischoff J, Arnold AE, Rojas E, Mejia LC, Woodward C, Kyllo DA. In: Biotic Interactions in the Tropics. Burslem DFRP, Pinard MA, Hartley SE, editors. Cambridge, UK: Cambridge Univ Press; 2005. pp. 226–237. [Google Scholar]
- 33.Davies TJ, Barraclough TG, Chase MW, Soltis PS, Soltis DE, Savolainen V. Proc Natl Acad Sci USA. 2004;101:1904–1909. doi: 10.1073/pnas.0308127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webb CO, Donoghue MJ. Mol Ecol Notes. 2005;5:181–183. [Google Scholar]
- 35.Hubbell SP, Foster RB, O'Brien ST, Harms KE, Condit R, Wechsler B, Wright SJ, Loo de Lao S. Science. 1999;283:554–557. doi: 10.1126/science.283.5401.554. [DOI] [PubMed] [Google Scholar]
- 36.Condit R. Tropical Forest Census Plots. Berlin: Springer; 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.