Abstract
The high-latitude planktonic foraminifera have proved to be particularly useful model organisms for the study of global patterns of vicariance and gene flow in the oceans. Such studies demonstrate that gene flow can occur over enormous distances in the pelagic marine environment leading to cosmopolitanism but also that there are ecological and geographical barriers to gene flow producing biogeographic structure. Here, we have undertaken a comprehensive global study of genetic diversity within a marine protist species, the high-latitude planktonic foraminiferan Neogloboquadrina pachyderma. We present extensive new data sets from the North Pacific and Arctic Oceans that, in combination with our earlier data from the North Atlantic and Southern Oceans, allow us to determine the global phylogeography of this species. The new genetic data reveal a pattern of Arctic circumpolar isolation and bipolar asymmetry between the Atlantic and Pacific Oceans. We show that the ancestry of North Pacific N. pachyderma is relatively recent. It lies within the upwelling systems and subpolar waters of the Southern Hemisphere and remarkably not within the neighboring Arctic Ocean. Instead, the Arctic Ocean population forms a genetic continuum with the North Atlantic population, which became isolated from the southern populations much earlier, after the onset of Northern hemisphere glaciation. Data from the planktonic foraminiferal morphospecies Globigerina bulloides is also introduced to highlight the isolation and endemism found within the subpolar North Pacific gyre. These data provide perspective for interpretation and discussion of global gene flow and speciation patterns in the plankton.
Keywords: global biogeography, Globigerina bulloides, Neogloboquadrina pachyderma, planktonic foraminifera
Molecular studies have revealed extensive genetic diversity in many apparently cosmopolitan species of marine planktonic protists (1). These genetically discrete but morphologically similar genotypes frequently exhibit unique biogeographies, distinct ecologies, and novel adaptations that are consistent with species-level classification (2–8). Clearly, skeletal morphology is not sufficient to distinguish microbial species, although in some cases, once difference is highlighted by genetic characterization, it has proved possible to detect subtle morphological differences in a planktonic foraminifer (4) and also in some coccolithophores (7) and diatoms (9–10).
A lack of morphological characters and difficulty in applying conventional species concepts has generated circuitous reasoning and often heated debate on the recognition of microbial biogeography and patterns of evolution (11–13). Characterization of biodiversity in microbes requires a combination of genetic, ecological, and biogeographical information (14), but to examine the evolutionary processes requires interpretations within a historical context (1, 15). Complete fossil records are limited to very few microbial groups, and the high-resolution planktonic foraminiferal fossil archive, in particular, provides unequalled historical perspective for such integrated studies. In addition, planktonic foraminifera exhibit an unusually high rate of evolution in their small-subunit ribosomal RNA (SSU rRNA) gene sequences, providing resolution to species level (16) that is rarely observed in other groups.
Studies using this integrated approach indicate that a broad range of evolutionary processes can cooccur, even within closely related groups of taxa in the pelagic environment (15). For example, SSU rRNA genotypes are identical in several planktonic foraminiferal species in the northern and southern Atlantic subpolar water masses (15), showing that the extensive tropics and subtropics of the Atlantic are not sufficiently hostile to prevent transtropical gene flow between subpolar-adapted marine protists. At first sight, this early phylogeographic evidence from planktonic foraminifers seemed to support a “ubiquity” dispersal scenario (11), where genetic types appear unconfined by geographical or oceanic boundaries (5, 15). However, recent evidence from the highest polar latitudes of the North Atlantic suggests that vicariant differentiation and allopatric processes prevail in the polar, more isolated regions (1). Reconstruction of the evolutionary processes driving the morphologically cryptic genetic divergences observed in the bipolar planktonic foraminiferal morphospecies Neogloboquadrina pachyderma show that this morphospecies has an antitropical polar distribution throughout its ecological range in the Atlantic, yet the sibling genetic types found within the polar provinces do not exhibit a “ubiquitous” distribution pattern (1).
N. pachyderma dominates planktonic foraminifer assemblages throughout the high-latitude marine provinces of both hemispheres. To obtain a global picture of the genetic diversity within this ubiquitous polar morphospecies, we have expanded our collections into the Arctic Ocean and North Pacific. Molecular genetic data from these regions are essential to determine whether the Northern Hemisphere populations form a genetic continuum and whether the interhemispheric isolation that occurred after the onset of Northern Hemisphere glaciation in the Atlantic (1) also happened in the Pacific. We have further increased our database of DNA sequences from the Benguela upwelling region in a bid to assemble the first truly global phylogeography and evolutionary history of a marine pelagic protist species. To provide a wider perspective for interpretation, we have introduced new data from the planktonic foraminiferal morphospecies Globigerina bulloides to explore the endemism of the North Pacific subpolar gyre and comparative patterns of gene flow in the Pacific and Atlantic Oceans.
Results
Biogeography of Arctic Subpolar and Polar N. pachyderma Genotypes.
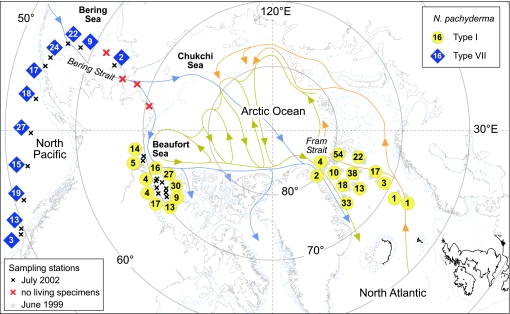
A total of 524 genotyped specimens from the North Pacific, Arctic Ocean, and North Atlantic were used in the study. Two N. pachyderma SSU genotypes were identified in the North Pacific and Beaufort Sea (Arctic Ocean) by either sequencing or restriction digest (see Materials and Methods). The North Pacific genotype (Type VII, n = 169) was distinct from all other types described to date (1). The Arctic Ocean specimens (n = 139) were found to be Type I. This genotype was previously identified in the polar North Atlantic (1) (n = 216; Fig. 1).
Fig. 1.
Biogeographical distribution of SSU rRNA genotypes of N. pachyderma throughout the North Pacific (Type VII, n = 169), Arctic (Type I, n = 139), and North Atlantic (Type I, n = 216). The North Atlantic data were collected in a previous study (1). The surface ocean currents (blue, North Pacific water; orange, North Atlantic water) and the Arctic Ocean intermediate layer circulation pattern (dark yellow) are shown (17). Specimen numbers at each station are indicated within the symbols for each genotype.
Limited variation was observed among the tandem repeats of the SSU rRNA gene within individual specimens of N. pachyderma. Tandem repeat variation is rare in planktonic foraminifer species and has only been detected in a limited number of groups, including the Neogloboquadrina and Globigerinita lineages evidenced from our databases. A survey of the extent of intraindividual variation in the first ≈500 base pairs of the SSU rRNA gene fragment for 155 representative clones from 19 individuals from the North Pacific, Arctic, and North Atlantic Oceans revealed a complete lack of commonality in the variant gene repeats between the North Pacific N. pachyderma Type VII and Arctic/North Atlantic N. pachyderma Type I genotypes, highlighting the absence of gene flow between these two populations.
The genotype N. pachyderma Type VII was found in the eastern North Pacific between Vancouver Island and the shelf edge in the Bering Sea with sea surface temperatures (SSTs) between 14°C and 9°C. Although there is a considerable flow of water from the North Pacific into the Arctic Ocean that would be expected to carry passively floating plankton north (18), no living planktonic foraminifers were found in the shallow region of the Bering Strait and Chukchi Sea over a distance of >1,000 km. In the Arctic Ocean, a prolific population of N. pachyderma Type I was found off the shelf edge of the Beaufort Sea in temperatures ranging from 10°C to 0.4°C. The Type I genotype had previously been found only in the North Atlantic, where it was found to be the sole genotype of N. pachyderma present in these waters (1). The Arctic Ocean N. pachyderma Type I population therefore forms a continuum with that of the North Atlantic Type I. The North Pacific N. pachyderma Type VII population, on the other hand, is genetically isolated, and there is no evidence of low-level expatriation of either genotype across the Bering gateway in the genotype assemblages.
G. bulloides Biogeography.
Specimens of a second species of planktonic foraminifera, G. bulloides, were also collected along the North Pacific transect shown in Fig. 1. A single genotype (Type IIe, n = 124) of this subpolar species was identified and was found to be highly distinct from all other genotypes of this species known to date (19–20). No other G. bulloides genotypes were found in the North Pacific subpolar gyre during our collection in the northern summer of 2002.
N pachyderma Phylogeny.
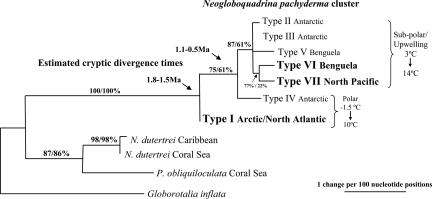
The evolutionary interrelationships among all N. pachyderma genotypes identified to date are shown in Fig. 2. The maximum-likelihood (ML) phylogeny includes new genotypes from the North Pacific (Type VII) and Benguela System (Type VI). It is consistent with phylogenies constructed by using neighbor-joining (NJ), Bayesian inference (BI), Fitch Margoliash (FM), and maximum parsimony (MP). All N. pachyderma genotypes cluster together with 100% NJ/ML bootstrap support, and the N. pachyderma lineage is highly distinct from the Neogloboquadrina dutertrei/Pulleniatina obliquiloculata lineage. The first major divergence in the N. pachyderma genotype cluster separates the polar North Atlantic and Arctic Ocean Type I from the remaining N. pachyderma genotypes. The second divergence separates the Southern Hemisphere polar genotype, Type IV (75%/61% NJ/ML bootstrap support) from the remaining subpolar/upwelling genotypes, which cluster together (87%/61% NJ/ML bootstrap support). This group includes the newly characterized North Pacific genotype (Type VII) and the new South Atlantic Benguela genotype (Type VI). N. pachyderma Type VI is the second genotype to be found within the Benguela System off Namibia and represents an additional potentially relict genotype within the Benguela System after isolation from Southern Ocean populations during interglacial periods (1). The genotypes within this subpolar and upwelling cluster are defined mainly by differences in the variable regions of the SSU rDNA sequence (1). However, there is a hint (77%/22% NJ/ML bootstrap support) that the new Benguela Type VI and the North Pacific N. pachyderma Type VII may have diverged from a common ancestor, although we note that the ML bootstrap is very low.
Fig. 2.
Maximum-likelihood phylogenetic tree showing the evolutionary relationships among the N. pachyderma genotypes. The tree is an extension of the phylogeny of the Neogloboquadrina group (1) to include previously undescribed genotypes from the North Pacific (Type VII), Arctic Ocean (Type I), and Benguela System (Type VI), which are highlighted in bold. The tree was constructed from 840 nucleotide sites of the SSU rDNA gene and is rooted on G. inflata. Cryptic divergence time estimates are shown (1). Subpolar/upwelling and polar adapted N. pachyderma genotypes are delineated. Bootstrap values (NJ/ML; expressed as a percentage) indicate support for branches in the tree.
G. bulloides Phylogeny.
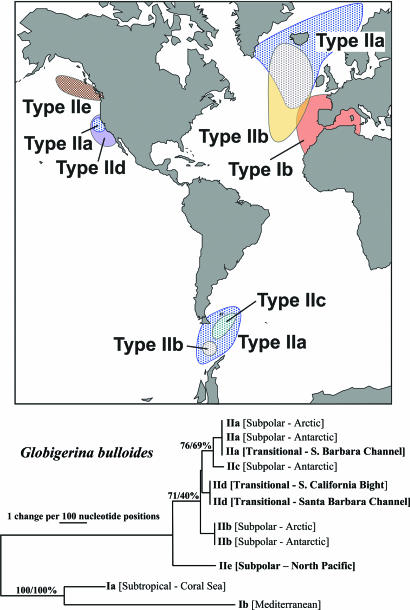
The evolutionary interrelationships among the G. bulloides genotypes identified to date are shown in Fig. 3. The ML phylogeny includes a new genotype from the North Pacific (Type IIe). It is consistent with phylogenies constructed by using NJ, BI, FM, and MP. The subtropical genotypes (Type I) consistently fall basal within the G. bulloides cluster in foraminiferal phylogenetic trees (15). Their early divergence makes them a suitable outgroup to examine the direction of evolution within the Type II cluster. There are five clear genetic types within the cool water Type II cluster (Types IIa–IIe), and the North Pacific subpolar G. bulloides Type IIe diverges first from the common ancestor of the group. Branch lengths suggest that this divergence is relatively early when compared with the divergences of the other Type II genotypes. The later divergences may be associated with Quaternary climate cyclicity (20) and are consistent with the Quaternary divergences observed in N. pachyderma (1).
Fig. 3.
Maximum-likelihood phylogenetic tree showing the evolutionary relationships among G. bulloides transitional and subpolar genotypes (20). The tree is based on analysis of 729 nucleotides of the SSU rRNA gene and is rooted on the subtropical G. bulloides Type I genotypes (20). Bootstrap values (NJ/ML; expressed as a percentage) indicate support for branches in the tree.
Discussion
Northern Hemisphere Isolation.
The North Atlantic/Arctic population.
The extensive sampling (n = 355) of N. pachyderma carried out throughout the North Atlantic and Arctic water mass indicates that only a single N. pachyderma genotype (Type I) frequents these waters. Molecular clock estimates suggest that the isolation of this genotype occurred within the early Quaternary as a result of Northern Hemisphere cooling, between 1.8 and 1.5 million years ago (Ma) (1) (Fig. 2). The cold-adapted N. pachyderma Type I genotype has been unable to transit the tropics since and has remained genetically and geographically isolated in the Arctic Ocean and North Atlantic until the present day. Ocean circulation patterns dictate that North Atlantic planktonic foraminifers would be carried into the Arctic Ocean circulation by the North Atlantic Current and exit through the Fram Strait via the East Greenland current (Fig. 1). Constant mixing of passively floating plankton in the Arctic circulation is consistent with the presence of a single genotype within these water masses.
Transarctic gene flow: tectonic and ecological constraints.
Given the high dispersal potential of the free-floating planktonic foraminifera and the existence of a transarctic marine connection via the Bering Strait, one might assume that the Northern Hemisphere populations of polar planktonic foraminifera should represent a single circumpolar species or that frequent genetic exchange across the Arctic should have taken place during sea level high stands. The opening of the Bering Strait in the latest Miocene (4.8–5.5 Ma) (21), provided a new high-latitude marine gateway between the North Pacific and North Atlantic. Large numbers of taxa migrated across the northern latitudes initially, with invaders from the North Pacific far outnumbering those from the Arctic–Atlantic (22). Current ocean circulation patterns dictate that passive planktonic dispersers would be carried by strong directional flow into the Arctic Ocean from the North Pacific (Fig. 1) (18), although glacioeustatic sea-level cyclicity periodically closes the shallow Bering Sea gateway. Molecular studies show that many Pacific marine organisms transit the Arctic sporadically during sea level high stands in interglacial periods. These include sea urchins (23), bivalves (24), and a red alga (25). Recent studies from circumArctic sea urchins even suggest that gene flow can be in both directions (26). Transiting taxa mostly have planktonic larvae as part of their life cycle, which facilitates passage. Marine planktonic protists would therefore be expected to be able to cross the Bering Strait because of their high dispersal potential, predominantly asexual reproduction strategies, and, often, the ability to form resistant spores.
The existence of two distinct and distantly related genetic types of N. pachyderma (Type I and Type VII, Fig. 2) with no evidence of hybridization clearly demonstrates that the above scenario is not the case for planktonic foraminifers. Planktonic foraminifers are relatively conservative in reproductive habit because they are currently thought to only reproduce sexually, by using various strategies to facilitate spatially and temporally synchronous release of gametes. Under such constraint, their life cycle would be successfully completed only in connection with a vertical migration down through the water column. The depth of the Bering and Chukchi Seas falls to <50 m, most likely precluding foraminiferal sexual reproduction in this region. Transit time within the shallow Bering corridor (18) may also be sufficiently long to exclude transfer of N. pachyderma colonizers, because it takes a much longer time than the expected life cycle of these high-latitude morphospecies (27). Planktonic foraminifera do not have an encystment stage, which would prolong their life cycle to aid passage through inhospitable regions.
N. pachyderma Type VII may also be confined to the subpolar North Pacific by its ecological adaptations. There is no true polar province in the North Pacific (28), and the newly characterized North Pacific N. pachyderma Type VII is thus a subpolar dweller. N. pachyderma Type I is the only genotype populating the whole of the North Atlantic and Arctic Ocean polar province (Fig. 1). Driven by progressive cooling in the Northern Hemisphere and the formation of the strong isolating Gulf Stream in the North Atlantic, N. pachyderma Type I is thought to have become transformed from a bipolar subpolar cosmopolitan into an Arctic polar specialist (1). It is confined to live at the coldest ocean temperatures between −1.6°C and 10°C and exposed to the rigors of sea ice cover in Northern winters. N. pachyderma Type I must have a strategy for overwintering in Arctic polar waters or else it would not have been able to repopulate open-water regions in the Canadian Arctic. In the Antarctic, N. pachyderma Type IV lives in polar waters below 2°C and is adapted to feed and grow while overwintering in the brine channels within sea ice (29). However, N. pachyderma Type I have never been observed inhabiting sea ice in the Arctic.
The N. pachyderma phylogeny suggests that both divergence and clustering within the tree are associated with identifiable barriers and ecological drivers and linkages (Fig. 2). Consistent with its subpolar status, N. pachyderma Type VII clusters with the other subpolar and upwelling genotypes of the Southern Hemisphere and not with the polar Northern Hemisphere North Atlantic/Arctic Type I (Fig. 2). Members of this group are not associated with subzero temperatures or seasonal sea ice and were found in SSTs ranging between 2°C and 14°C. Sediment trap data from the subpolar northwest Pacific (30) show that the ecological preferences of N. pachyderma from the region are consistent with those of the subpolar Southern Ocean genotypes II and III (1). In the northwestern Pacific, N. pachyderma are found throughout the year in SST's ranging between 2°C and 12°C with maximal numbers associated with temperatures of 4.8°C, high surface nutrient concentrations, and low stratification (30).
Transequatorial Relationships.
Although North Pacific N. pachyderma Type VII clusters with the Southern Ocean subpolar genotypes, there is no evidence from the variable regions of their SSU sequences that they currently intermix. What is highly surprising is that the divergence of the North Pacific genotype from the Southern Ocean genotypes must have occurred much more recently than the divergence between the North Atlantic and Southern Ocean genotypes (Fig. 2), which was dated at between 1.8 and 1.5 Ma (1). The close relationship between Type VII and the Benguela upwelling Type VI hints that the North Pacific N. pachyderma may have diverged from a common ancestor in the Southern Ocean in the late Quaternary. There are two scenarios to explain the origin of the North Pacific genotype. First, it may represent a vicariant population that became isolated from the Southern Ocean populations, most likely as a result of perturbations in the Quaternary climate. This would imply that the ancestor of the currently isolated North Pacific Type VII was originally bipolar and able to maintain genetic exchange across the tropics. Alternatively, it may be the result of the dispersal of a Southern Ocean genotype into the North Pacific. This scenario would require both a dispersal event from the Southern Ocean and the extinction of the original resident N. pachyderma in the North Pacific. Both scenarios require the establishment of a permanent dispersal barrier preventing other Southern Ocean Types from crossing the tropical Pacific at some time in the late Quaternary. Neither scenario excludes the possibility that other isolated N. pachyderma genotypes may occur in the western region of the North Pacific subpolar water mass.
Endemism in the North Pacific.
The barriers to antitropicality in the Pacific appear less formidable than in the Atlantic. The subpolar provinces impinge directly onto the equatorial provinces in the Pacific, and the South American upwelling regime extends very close to the equator, potentially facilitating exchange of temperate species between the hemispheres (28). This, together with the subpolar rather than polar adaptation of the Southern Ocean N. pachyderma Type II and III and North Pacific Type VII, may explain why the isolation of the North Pacific N. pachyderma occurred substantially later than in the Atlantic, where the extensive subtropical provinces isolate the subpolar provinces by considerable distances. The isolation of Type VII in the North Pacific occurred after the Antarctic Type IV adaptive divergence to overwinter in the sea ice, dated at between 1.1 and 0.5 Ma and thought to be linked to the onset of the late Quaternary glacial–interglacial cyclicity (1). The isolation of the North Pacific Type VII must have occurred before the current interglacial, because we see no evidence for genetic exchange with the Southern Ocean in the present day in N. pachyderma.
North Pacific isolation is not unique to N. pachyderma. Although the same subpolar planktonic foraminiferal morphospecies with antitropical distributions occur in the Pacific as in the Atlantic, data from the California Margin (20) and off Vancouver Island (31) indicate that planktonic foraminiferal endemism is prevalent throughout the transitional and subpolar water masses. The California Margin G. bulloides Type IId and Neogloboquadrina incompta Type II (20, 31) are genetically distinct from those in the Southern Ocean. These genotypes have not been found in any other water mass to date. In this study, we present further evidence of the extensive endemism prevailing in the North Pacific subpolar gyre. The newly described genotype, G. bulloides Type IIe (Fig. 3), collected at the sampling stations shown in Fig. 1, was the sole G. bulloides genotype found in these waters. Only a single specimen of G. bulloides Type IIa has been identified to date in the North Pacific off the Californian Margin (20), which is common to the subpolar regions of the North and South Atlantic. The transient appearance of this genotype in January was explained as a seasonal incursion of the cool California Current into the Santa Barbara Channel, with the assumption that the subpolar G. bulloides assemblage in the North Pacific mirrored that of the North Atlantic. The evidence presented here indicates that G. bulloides Type IIe is the sole subpolar genotype frequenting the North Pacific gyre and not the bipolar genotypes Types IIa and IIb as in the North and South Atlantic (Fig. 3 and ref. 15). The single G. bulloides Type IIa specimen found in the Santa Barbara Channel (20) must have come either from the western North Pacific or from the south.
The newly identified G. bulloides Type IIe constitutes the earliest divergence in the cool water genotype cluster (Fig. 3). All other G. bulloides genotypes, including Type IId from the California Margin, share a common ancestor. The basal position of Type IIe within the cool water cluster possibly hints that the cool water types could have arisen in the North Pacific and radiated from there. North Pacific endemism in planktonic foraminifers is highly consistent with many other taxonomic groups and the North Pacific may function as an important center of origin (32) of successful foraminiferal species. The North Pacific has a high level of species diversity compared with the North Atlantic, which is thought to be due to its larger size, heat budget, and moderated temperature fluctuations being sheltered by the protective Bering land bridge (32). However, this does not explain why we do not see bipolar genotypes within the North Pacific. Subpolar planktonic foraminifera are quite capable of transiting the equatorial regions of the Atlantic (15), so why not then the Pacific? There is clearly a barrier to gene flow northwards, which, from evidence presented here for the eastern North Pacific, is breached very rarely, such as in the case of N. pachyderma Type VII. The North Pacific biota is thought to resist penetration from other areas where incoming species may be less competitive or less fit (32). This may be applicable to pelagic protists also and may indicate that the North Pacific genotypes will have different ecologies from their sibling counterparts. Whether the North Pacific barrier to incoming planktonic foraminiferal genotypes is confined to the subarctic water mass remains to be determined.
Evolutionary Processes.
The phylobiogeographic data presented here provide a global overview of the distribution pattern of morphologically cryptic genetic types within a marine pelagic protist. The chronology of the isolation of N. pachyderma in the Pacific Ocean does not mirror that of the Atlantic, and the evolutionary processes act independently of one another in each ocean. The isolated North Atlantic N. pachyderma Type I forms a genetic continuum throughout the whole of the northern Arctic polar province. The North Pacific genotype, on the other hand, became isolated at a much later period within the Quaternary and has closer evolutionary links with the Southern Ocean. The North Pacific subpolar province also harbors endemic genotypes of other subpolar morphospecies, which is not observed in the North Atlantic.
It is possible to argue that, before the Quaternary. N. pachyderma could have constituted a globally dispersed bipolar metapopulation (11) as all extant cryptic genotypes diverged from a common ancestor. However, their morphologically cryptic character provides no clues to lineage extinction, which could have occurred at any time since their appearance 10.4 Ma ago (1) and leaves their past potential ubiquity forever in question. The recently evolved genotypes have distinct biogeographies and different adaptations, and evidence indicates that there are several co-occurring evolutionary mechanisms responsible for their recent radiation. Already, there is strong evidence that high-dispersal marine protists such as planktonic foraminifers exhibit both regional cosmopolitan type distributions and vicariant geographical isolation in the Atlantic (1, 15). Our data from the North Pacific and Arctic indicate that vicariant isolation predominates in the subpolar North Pacific and that a locally ubiquitous population prevails within the Arctic Ocean and North Atlantic. The Bering Sea gateway is closed to N. pachyderma, creating a physical barrier that must have always isolated the Arctic Ocean N. pachyderma population from that of the North Pacific. Instead, gene flow in the North Pacific population is restricted to intermittent mixing with the very distant subpolar Antarctic populations in the later Quaternary. The isolation between the North and South Pacific populations is much later than the isolation observed between the North and South Atlantic N. pachyderma in the early Quaternary, estimated at 1.8–1.5 Ma (1). Such asymmetry in transtropical gene flow must reflect the disparity in hydrographic character of the Atlantic and Pacific oceans. Disparity in gene flow is also reinforced by the high degree of endemism within the North Pacific.
The ubiquity hypothesis (11) is constrained by the degree of resilience of dispersing stages. Clearly, planktonic foraminifers do not always have unlimited dispersal ability. They are constrained by physical barriers such as the shallow Bering and Chukchi Seas (this study) and the African Cape (31), which dissects their provincial domains and prevents passage over long periods of time. Any rare expatriations would carry them into domains to which they are not adapted to live. In addition, they are constrained by oceanographic barriers such as the tropics and subtropics (ref. 1 and this study) and frontal systems (1), where ecological processes are additionally recruited and play an increasingly important isolating role. Ecological constraints thus appear to be major drivers of divergence in the pelagic realm.
Materials and Methods
Sampling Localities.
N. pachyderma specimens (Type VII and Type I) from the north Eastern Pacific, Bering Sea, and Arctic Ocean were collected onboard the Canadian Coast Guard Ship Sir Wilfrid Laurier (2002) either by pumping water samples from the ship's nontoxic water supply (6-m depth, 83-μm mesh) or by vertical net tows (100-m depth, 83-μm mesh) along a transect from Vancouver Island in the Eastern Pacific (49°23′N/127°40′W) to the Beaufort Sea in the Arctic Ocean (69°54′N/121°21′W). In the previous study in the Arctic Atlantic, N. pachyderma (Type I) specimens were collected onboard RV Polarstern (ARKXV/1 + 2, 1999). Specimens were obtained by using multinets (upper 500 m, 63-μm mesh) at 75°N (from 13°W to 13°E) and from a mixture of multinet and surface pumped water samples (6-m depth, 63-μm mesh) in the Nordic seas between 80°N and 60°N (1, 15). N. pachyderma samples from the Benguela system off Namibia were collected from RV Welwitschia (November 2001) by vertical plankton tow (50 m, 63-μm mesh) along a cruise transect between 2 and 30 nautical miles from the coast at 23°S (1). The additional N. pachyderma genotypes (II, III, and IV) were collected from different water masses in the Southern Ocean (1). G. bulloides Type IIe specimens were also collected from the north Eastern Pacific and Bering Sea onboard the Canadian Coast Guard Ship Sir Wilfrid Laurier (2002) either by pumping water samples from the ship's nontoxic water supply or by vertical net tows along a transect from Vancouver Island in the Eastern Pacific (49°23′N/127°40′W) to (58° 13′N/173°19′W).
Isolation and Sequencing of SSU Genes.
DNA extraction, amplification by PCR, and automated sequencing of ≈1,000-bp and ≈500-bp fragments of the terminal 3′ end of the foraminiferal SSU rRNA gene were as described previously (1, 31). Because of a limited but defined degree of ambiguity, complete fragment direct sequencing was not possible for all specimens. PCR products from 19 individuals were therefore cloned by using a PCR 2.1 TOPO TA cloning kit (Invitrogen, Carlsbad, CA), and 155 clones were amplified. From the analysis of these sequences, a restriction digestion strategy was devised by using DraI. The digest produces fragment patterns which unequivocally differentiate Type I from Type VII. Type I specimens were partially sequenced to confirm their correct genotype assignment.
Phylogenetic Analysis.
Partial SSU rDNA sequences were aligned manually within version 2.2 of the Genetic Data Environment (GDE) package (33). Phylogenetic trees for N. pachyderma were based on 840 unambiguously aligned nucleotide sites and rooted on the globorotaliid Globorotalia inflata. For G. bulloides, phylogenies were based on 729 sites and rooted on the subtropical G. bulloides Type I genotypes. Phylogenetic trees were constructed by using the NJ, FM, ML, and MP methods within PAUP* version 4.0d64 (34). For the NJ, FM, and ML methods a general time reversible (GTR) model was used, with rate heterogeneity between sites accounted for by incorporating γ-distributed rates in the model (Γ). The rate matrix, base frequencies, and shape parameter (α) of the γ-distribution (based on 16 rate categories) were estimated by using likelihood by iteration from an initial NJ tree. Genetic distances were estimated by using the GTR+Γ model. Bootstrap resampling (1,000 replicates) was used to assign support to branches in the trees (35). BI was performed by using the MrBayes (version 3.0B4) package (36). A GTR+Γ model (16 rate categories) was used and the tree space was explored by using four chains of a Markov Chain Monte Carlo algorithm for 1 million generations, sampling every 100 generations. A consensus tree was built by using the last 1,000 trees (burnin = 9,001 samples). The sequences in this study are deposited in GenBank under the accession nos. EF447102–EF447104.
Acknowledgments
We thank Bon van Hardenberg and the Canadian Coast Guard and crew for logistical and technical assistance during sampling on the CCGS Sir Wilfrid Laurier, Fiona McLaughlin and Eddy Carmack for facilitating our participation on the CCGS Sir Wilfrid Laurier cruise to the Canadian Arctic in 2002, and Fred Kippert for molecular advice and assistance. We gratefully appreciate the support of the Research Institute of the Ministry of Fisheries and Resources (Namibia) staff for sampling from RV Welwitschia. This work was supported by Natural Environment Research Council of the United Kingdom Grants NER/J/S/2000/00860 and NER/M/S/2001/00476.
Abbreviations
- SSU
small subunit
- ML
maximum likelihood
- NJ
neighbor joining
- GTR
general time reversible.
Footnotes
References
- 1.Darling KF, Kucera M, Pudsey CJ, Wade CM. Proc Natl Acad Sci USA. 2004;101:7657–7662. doi: 10.1073/pnas.0402401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knowlton N. Annu Rev Ecol Syst. 1993;24:189–216. [Google Scholar]
- 3.Knowlton N. Hydrobiologia. 2000;420:73–90. [Google Scholar]
- 4.Huber BT, Bijma J, Darling KF. Paleobiology. 1997;23:33–62. [Google Scholar]
- 5.Darling KF, Wade CM, Kroon D, Leigh Brown AJ, Bijma J. Paleoceanography. 1999;14:3–12. [Google Scholar]
- 6.de Vargas C, Norris R, Zaninetti L, Gibb SW, Pawlowski J. Proc Natl Acad Sci USA. 1999;96:2864–2868. doi: 10.1073/pnas.96.6.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saez AG, Probert I, Geisen M, Quinn P, Young JR, Medlin LK. Proc Natl Acad Sci USA. 2003;100:7163–7168. doi: 10.1073/pnas.1132069100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uwe J, Fensome RA, Medlin LK. Mol Biol Evol. 2003;20:1015–1024. doi: 10.1093/molbev/msg105. [DOI] [PubMed] [Google Scholar]
- 9.Mann DG, McDonald SM, Bayer MM, Droop SJM, Chepurnov VA, Loke RE, Ciobanu A, du Buf JMH. Phycologia. 2004;43:459–482. [Google Scholar]
- 10.Sarno D, Kooistra WHCF, Medlin LK, Percopo I, Zingone A. J Phycol. 2005;41:151–176. [Google Scholar]
- 11.Finlay BJ. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 12.Fenchel T. Aquat Microb Ecol. 2005;41:49–54. [Google Scholar]
- 13.Mitchell EAD, Meisterfeld R. Protist. 2005;156:263–267. doi: 10.1016/j.protis.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Finlay BJ. Philos Trans R Soc London B. 2004;359:599–610. doi: 10.1098/rstb.2003.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darling KF, Wade CM, Steward IA, Kroon D, Dingle R, Leigh Brown AJ. Nature. 2000;405:43–47. doi: 10.1038/35011002. [DOI] [PubMed] [Google Scholar]
- 16.Pawlowski J, Bolivar I, Fahrni J, de Vargas C, Gouy M, Zaninetti L. Mol Biol Evol. 1997;14:498–505. doi: 10.1093/oxfordjournals.molbev.a025786. [DOI] [PubMed] [Google Scholar]
- 17.Jones EP. Polar Res. 2001;20:139–146. [Google Scholar]
- 18.Woodgate R, Aagaard AK, Weingartner TJ. Deep Sea Res Part II. 2005;52:3116–3149. [Google Scholar]
- 19.Kucera M, Darling KF. Philos Trans R Soc London A. 2002;360:695–719. doi: 10.1098/rsta.2001.0962. [DOI] [PubMed] [Google Scholar]
- 20.Darling KF, Kucera M, Wade C, von Langen P, Pak D. Paleoceanography. 2003;18:1032. [Google Scholar]
- 21.Marincovich L, Gladenkov AY. Nature. 1999;397:149–151. [Google Scholar]
- 22.Vermeij GJ. Paleobiology. 1991;17:281–307. [Google Scholar]
- 23.Palumbi SR, Kessing BD. Evolution (Lawrence, Kans) 1991;45:1790–1805. doi: 10.1111/j.1558-5646.1991.tb02688.x. [DOI] [PubMed] [Google Scholar]
- 24.Väinölä R. Mar Biol. 2003;143:935–946. [Google Scholar]
- 25.Van Oppen MJH, Draisma SG, Olsen JL, Stam WT. Mar Biol. 1995;123:179–188. [Google Scholar]
- 26.Addison JA, Hart MW. Evolution(Lawrence, Kans) 2005;59:532–543. [Google Scholar]
- 27.Schiebel R, Hemleben C. Paläontologische Zeitschrift. 2005;79:135–148. [Google Scholar]
- 28.Lipps JH. Palaeogeogr Palaeoclimatol Palaeoecol. 1972;12:3–14. [Google Scholar]
- 29.Dieckmann GS, Spindler M, Lange MA, Ackley SF, Eicken H. J Foram Res. 1991;21:182–189. [Google Scholar]
- 30.Kuroyanagi A, Kawahata H, Nishi H, Honda MC. Deep Sea Res Part II. 2002;49:5627–5645. [Google Scholar]
- 31.Darling KF, Kucera M, Kroon D, Wade CM. Paleoceanography. 2006;21:PA2011. [Google Scholar]
- 32.Briggs JC. J Biogeogr. 2003;30:1–18. [Google Scholar]
- 33.Smith SW, Overbeek R, Woese CR, Gilbert W, Gillevet PM. Comput Appl Biosci. 1994;10:671–675. doi: 10.1093/bioinformatics/10.6.671. [DOI] [PubMed] [Google Scholar]
- 34.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2003. Version 4. [Google Scholar]
- 35.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 36.Huelsenbeck JP, Ronquist F. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]