Abstract
Mammalian centromeres are defined epigenetically. Although the physical nature of the epigenetic mark is unknown, nucleosomes in which CENP-A replaces histone H3 are at the foundation of centromeric chromatin. Hydrogen/deuterium exchange MS is now used to show that assembly into nucleosomes imposes stringent conformational constraints, reducing solvent accessibility in almost all histone regions by >3 orders of magnitude. Despite this, nucleosomes assembled with CENP-A are substantially more conformationally rigid than those assembled with histone H3 independent of DNA template. Substitution of the CENP-A centromere targeting domain into histone H3 to convert it into a centromere-targeted histone that can functionally replace CENP-A in centromere maintenance generates the same more rigid nucleosome, as does CENP-A. Thus, the targeting information directing CENP-A deposition at the centromere produces a structurally distinct nucleosome, supporting a CENP-A-driven self-assembly mechanism that mediates maintenance of centromere identity.
Keywords: centromere, chromatin, hydrogen exchange
Epigenetic determinants are known to modulate eukaryotic gene expression at the level of individual genes or entire chromosomes (1). Mounting evidence has implicated a similar epigenetic mechanism for the specification of the centromere, the chromosome element essential for faithful chromosome inheritance (2, 3). The centromere in metazoans is typically located within a region of repetitive satellite DNA in diverse plant and animal phyla (2, 4). In humans, the predominant centromeric satellite, αI, consists of repeats of 171-bp monomers that extend for several megabases at most centromeres (5). Despite the strong correlation between centromere location and the presence of these satellites, rare chromosomal rearrangements in humans have revealed instances in which a centromere has been silenced (6, 7) or generated de novo at a chromosome arm locus lacking detectable α satellite DNA (8–10) or both (11, 12). Such evidence strongly argues that centromere specification is not determined by a particular DNA sequence but rather is specified epigenetically (2, 3, 13).
The maintenance of centromere identity is critical because the loss of a single centromere or the presence of multiple, independently functioning centromeres on a chromosome will lead to a catastrophic cell division. In the former case, chromosome missegregation will lead to the loss of a specific chromosome in one daughter cell and an extra copy of it in the other. In the latter case, the chromosome will be subject to breakage by opposing forces exerted by the spindle. If centromere identity is lost in meiotic divisions, the gain or loss of a chromosome (aneuploidy) in the resulting gametes directly leads to spontaneous abortion or developmental defects in the resulting embryo. Mitotic aberrations leading to aneuploidy in dividing cells may contribute to cancer progression (14, 15), indicating that centromere identity in somatic cell lineages is of critical importance as well. All of this has raised the critical question: What determines the location of the centromere?
The most attractive candidate for an epigenetic mark that specifies the centromere is CENP-A, a histone variant that replaces H3 in centromeric nucleosomes in humans (16, 17), and CENP-A relatives have an essential role at centromeres in diverse eukaryotic species (2). CENP-A is found at all active centromeres in a manner that appears to be independent of DNA sequence, including human neocentromeres lacking detectable α satellites (10, 11). Conversely, despite the retention of αI-satellite arrays, CENP-A is absent when centromeres are silenced (10, 11). Nucleosomes assembled with CENP-A directly recruit a CENP-A nucleosome-associated complex (CENP-ANAC) that in turn recruits a set of at least seven more distal components (18). All of this has suggested that CENP-A-containing nucleosomes are an epigenetic mark for specifying location of the centromere (2, 13).
Here we have assembled nucleosomes with CENP-A and used hydrogen/deuterium (H/D) exchange coupled to MS (which measures amide proton exchange along the polypeptide backbone of each histone subunit) to determine that the CENP-A-containing nucleosome is more conformationally rigid than its canonical counterpart containing histone H3. Furthermore, this structural alteration is independent of DNA sequence but is driven by the CENP-A centromere targeting domain, or CATD (19), that can convert histone H3 into a centromere-targeted histone that can functionally replace CENP-A in centromere maintenance (20). These findings provide a structural basis for an epigenetic mark carried by CENP-A, and that is used to specify the location of the centromere.
Results
Protection from H/D Exchange Upon Assembly of Histones into Nucleosomes.
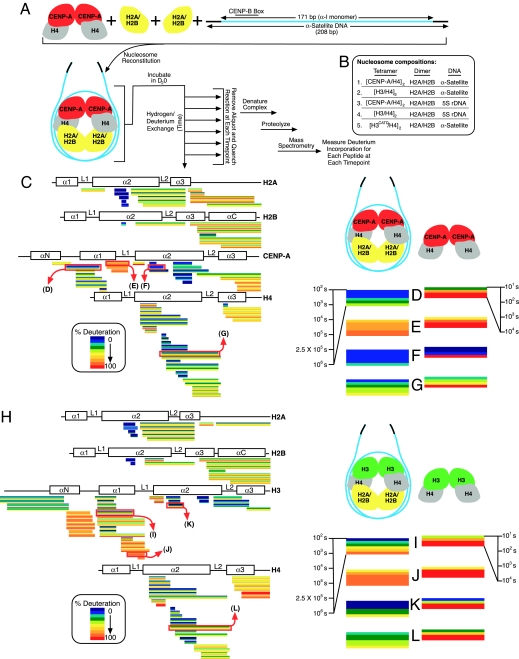
In static crystal structures, >3,000 water molecules are positioned within the canonical nucleosome structure and participate in the electrostatic interactions between histone subunits and between histones and DNA (21). The extent to which these water molecules are free to exchange in solution is not known. Centromeric or canonical nucleosomes were reconstituted by using the salt-dialysis method (22) starting with (CENP-A/H4)2 or (H3/H4)2 heterotetramers, H2A/H2B heterodimers, and a 208-bp DNA fragment encompassing a monomeric 171-bp human αI-satellite repeat (Fig. 1A). These nucleosomes were then incubated for times varying >4 orders of magnitude (102 to 106 s) in heavy water (D2O) to exchange deuterium onto amide protons along the polypeptide backbone. At each time point, exchange was quenched and the histones were then fragmented by proteolysis. Protection of the nucleosomal histones from rapid H/D exchange will occur if solvent does not have access for exchange (23). Slowed exchange also reflects increased stability of hydrogen bonding of amide protons. Protection of amide proton exchange can occur via intra- or intermolecular contacts, restricted conformational flexibility, compaction, or a combination of these.
Fig. 1.
Reduced H/D exchange upon assembly of histones into nucleosomes. (A) Experimental scheme for examining the solvent accessibility to nucleosomes assembled with αI-satellite DNA from human centromeres and with CENP-A in place of histone H3. (B) Composition of the five different nucleosomes reconstituted and examined by H/D exchange in this study. Either nucleosomes containing CENP-A (C–G) or histone H3.1 (H–L) were analyzed by H/D exchange. Horizontal blocks represent peptides from CENP-A-containing nucleosomes (C) or from H3-containing nucleosomes (H) monitored for H/D exchange over a time course from 102 to 106 s. Within each block the color-coded percentage of deuteration is represented with early time points at the top progressing to the latest time point at the bottom. Peptides are placed beneath schematics showing the location of the α-helices of each histone. Multiple charge states were detected for a subset of peptides, each represented by its own block. Peptides highlighted in C and H from the nucleosomes are enlarged (D–G Left and I–L Left) and compared with data collected from the corresponding subnucleosomal heterotetramers (D–G Right and I–L Right). The time course for the heterotetramer experiment extends over a range from 101 to 104 s, and these data are from published experiments (19).
Multiple peptides corresponding to the majority of each of the structured histone fold domains for each of the four histones of the 245-kDa nucleosome were then identified by MS, as well as the proportion of deuterium incorporated. Nucleosome assembly sharply reduced conformational accessibility within the folded cores of both H3 and H4 in the canonical nucleosome and CENP-A and H4 in the centromeric nucleosome, with most domains reduced at most sites by >3 orders of magnitude relative to the prenucleosomal heterotetramers (19) of either (H3/H4)2 or (CENP-A/H4)2. Thus, conformational flexibility of histones H3 (or CENP-A) and H4 are severely restricted by their incorporation into nucleosomes.
Close inspection of the CENP-A-containing nucleosomes revealed that some portions of the nucleosomal histone octamer, such as the region covered by the group of peptides spanning the αC-helix of histone H2B, showed little exchange at early time points, but nearly complete exchange later (Fig. 1C). Other regions, such as the α2-helix from CENP-A (Fig. 1 C and F), were protected from H/D exchange even after 106 s (≈12 days). Although this region was previously found to be the most conformationally constrained region of CENP-A in the subnucleosomal (CENP-A/H4)2 heterotetramer, complete exchange of this region in the heterotetramers was seen by 104 s (Fig. 1F) (19), whereas there was essentially no exchange in the CENP-A nucleosome even at 100-fold greater times (Fig. 1F), demonstrating that in the nucleosome this domain of CENP-A is held in a very rigid structure that does not undergo significant conformational flexing that would expose it to solvent. This offers direct support for the prediction, based on the known structure of the canonical nucleosome (24), that the majority of the α2-helix is buried within the center of the octameric disk-shaped core of the nucleosome as opposed to regions providing the major DNA contact points on the surface of the histone octamer.
CENP-A Directs the Formation of More Rigidified Nucleosomes.
The H/D exchange profiles of the CENP-A and H3 nucleosomes were similar, with most peptides reflecting a pattern of slow and fast exchanging regions in the CENP-A nucleosome that mimicked that seen for nucleosomes assembled with canonical histone H3 (H3.1) (Fig. 1H), consistent with the notion that CENP-A typically replaces histone H3 in a nucleosome with otherwise identical histone stoichiometries (two copies of each histone subunit) (17) and α-helical protein folds that are largely similar (Fig. 1 C and H).
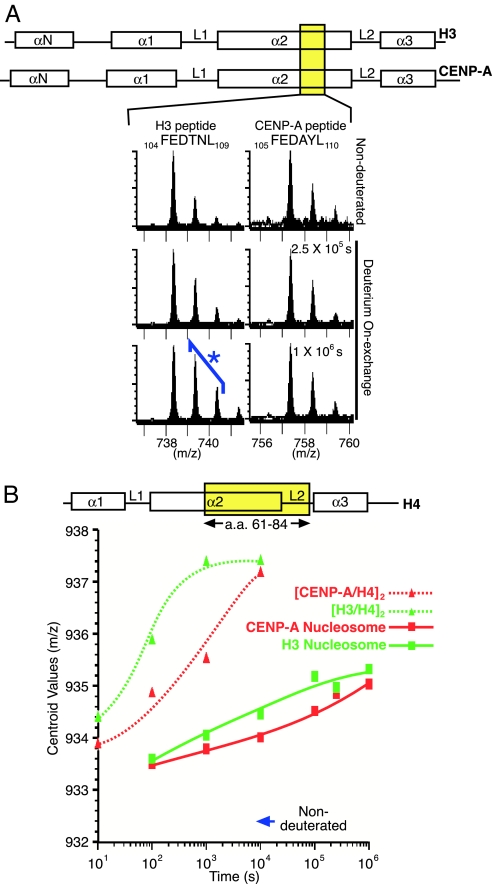
Histone H2A and H2B peptides from both CENP-A- and H3-containing nucleosomes displayed nearly identical exchange behavior. On the other hand, peptides from discrete regions of the centromeric nucleosome, including the α2-helix from CENP-A and the α2- and α3-helices from histone H4 were slower to exchange than the corresponding regions from the canonical nucleosome (Fig. 1 D–G and I–L). Although primary sequence differences between histone H3 and CENP-A limited comparison of peptides of identical composition, the residual sequence identity (the histone fold domains from H3 and CENP-A are 62% identical) allowed analysis of peptides that could potentially span regions of similar sequence. The FEDTNL peptide in H3 (amino acids 104–109) and FEDAYL peptide in CENP-A (amino acids 105–110) is one such peptide pair spanning the corresponding region within each respective α2-helix (Fig. 2A). Whereas measurable H/D exchange was observed at the 2.5 × 105 and 1 × 106 s time points for the H3 peptide (Fig. 2A), there was negligible exchange observed in the corresponding CENP-A peptide at all time points.
Fig. 2.
Centromeric nucleosomes containing CENP-A are more protected from H/D exchange relative to those containing histone H3. (A) Comparison of peptides from the corresponding position in the α2-helices from histone H3 and CENP-A. (B) Comparison of H/D exchange for an identical peptide from histone H4 (amino acids 61–84) in nucleosomes (assembled with either CENP-A or H3) or their respective subnucleosomal heterotetramers [(CENP-A/H4)2 or (H3/H4)2] (19).
Direct comparisons are possible when identical peptides are found corresponding to the histone subunits common to each type of nucleosome. From histone H4, a peptide spanning a large portion of the α2-helix was seen to exchange more slowly in the CENP-A nucleosome than when assembled into its canonical counterpart containing histone H3 [Fig. 2B and supporting information (SI) Fig. 5]. The α3-helix of histone H4 was also slower to exchange in the CENP-A-containing nucleosome, and this was evident even though the peptides identified to span the majority of this helix also contained the extreme C terminus including the rapidly exchanging, unstructured eight residues beyond the final α3-helix (19, 24). A second, slower phase of exchange was observed between this initial burst of exchange measurable at 1 × 102 s and the final time point (1 × 106 s) (SI Fig. 6). Thus, for each the α2- and α3-helices, the H4 peptides have undergone more H/D exchange at all time points in the canonical nucleosomes, whereas these same peptides require ≈10 times as long to achieve the same level of deuteration in the centromeric nucleosome.
Rigidity of the CENP-A-Containing Nucleosome Is Independent of DNA Sequence.
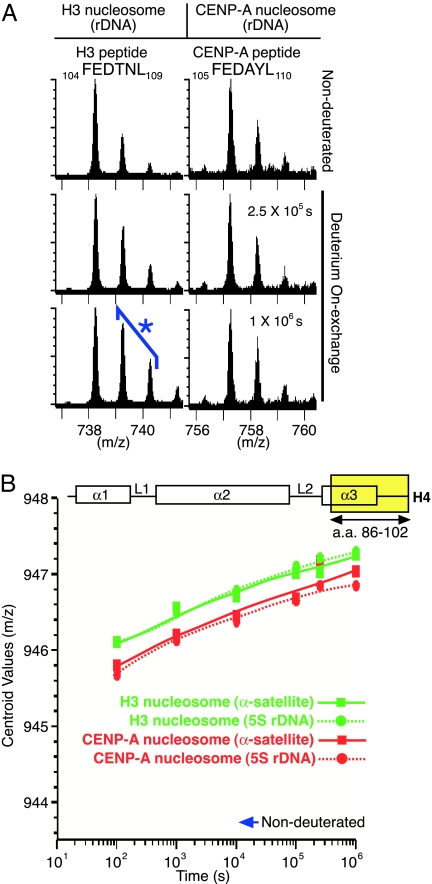
CENP-A-containing nucleosomes are typically assembled onto αI-satellite repeats in human chromosomes (17, 25), but functional neocentromeres that form at sites on chromosome arms lacking these repeats are loaded with CENP-A nonetheless (10, 11). To test whether the rigidity of the CENP-A nucleosome (Figs. 1 and 2 and SI Figs. 5 and 6) was selective to assembly with αI-satellite DNA, the H/D exchange behavior of CENP-A-containing and H3-containing nucleosomes was determined with a noncentromeric rDNA template (Fig. 3). Although there were some differences in the identities of the peptides recovered by using this rDNA (SI Figs. 7 and 8), the overall patterns of exchange were nearly identical. Moreover, closer inspection of similar (Fig. 3A) or identical (Fig. 3B and SI Figs. 9 and 10) peptides in the regions of rigidity of the CENP-A nucleosome revealed H/D exchange that was essentially indistinguishable when comparing wrapping with α-satellite or rDNA templates, indicating that conformationally more rigid nucleosomes are generated by CENP-A, rather than by influences from DNA sequences prevalent at typical centromeres.
Fig. 3.
The rigidified nucleosome structure generated by the incorporation of CENP-A does not require centromeric DNA. (A) The peptides (the same region as in Fig. 2A) are shown here from experiments in which the CENP-A-containing and H3-containing nucleosomes are now each assembled with rDNA. (B) Comparison of H/D exchange from an identical peptide from histone H4 from CENP-A-containing and H3-containing nucleosomes assembled with α-satellite DNA or rDNA.
The CATD Specifies Conformational Rigidity to Centromeric Nucleosome.
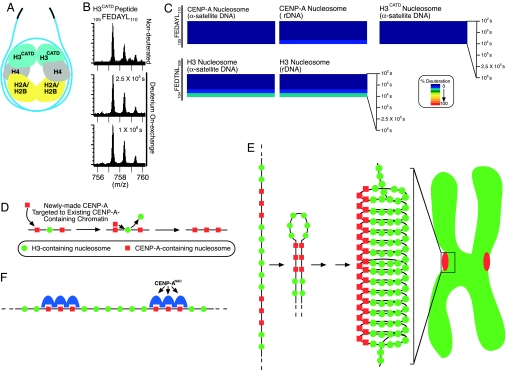
The conformationally more constrained subnucleosomal heterotetramer of (CENP-A/H4)2 and the targeting of newly made copies of this complex to existing centromeres has been shown to be determined by its CATD [this domain is formed by 22-aa changes from histone H3 within the Loop 1 (L1) and the α2-helix] (19). Substitution of the CATD into H3 is sufficient to replace most CENP-A in maintaining centromere function (20). To test whether the CATD plays a dual role serving both as a cis-acting targeting element on newly made (CENP-A/H4)2 heterotetramers en route to the centromere and as a chromatin-bound mark that generates structurally divergent nucleosomes unique to centromeres, nucleosomes were assembled with H3CATD, a chimeric version of histone H3 substituted with the CATD (19). The conformational rigidity of these nucleosomes was then determined by H/D exchange. The exchange in the H3CATD-containing nucleosome (Fig. 4A and SI Fig. 11) closely resembled that observed in bona fide centromeric nucleosomes assembled with CENP-A (Fig. 1C), with the L1 and α2-helix that constitute the CATD showing the very slow accessibility previously seen for the domain in CENP-A.
Fig. 4.
The CATD confers rigidity to the nucleosome. (A) Diagram of the H3CATD-containing nucleosome. (B) Negligible exchange is observed in the 105FEDAYL110 peptide from the CATD in the H3CATD nucleosome. (C) H/D exchange data corresponding to the FEDAYL peptide from CENP-A and H3CATD or FEDTNL from the H3-containing nucleosomes are plotted as described in Fig. 1. (D–F) Models for self-directed assembly of CENP-A nucleosomes (D), higher-order chromatin organization at the centromere driven by self-association of CENP-A-containing nucleosomes (E), and recruitment of the CENP-ANAC (18) via direct interaction with CENP-A-containing nucleosomes (F). See Discussion for details.
Close inspection of individual peptides that were found in peptide pools from both CENP-A- and H3CATD-containing nucleosomes showed a similar pattern of much slower exchange within a more conformationally constrained α2-helix (Fig. 4 B and C and SI Fig. 12) relative to histone H3-containing nucleosomes. Included here was an H3CATD peptide, corresponding to CENP-A amino acids 105–110 that showed almost no exchange throughout the entire time course. This behavior is nearly identical to that seen in the exact same peptide in the context of the CENP-A-containing nucleosome, but differed from the more readily exchangeable nature of the peptide at the corresponding position in the H3-containing nucleosome (amino acids 104–109) (Figs. 2A and 4C). Importantly, examination of peptide behavior from within the α2-helix of H4 revealed that incorporation of H3CATD into a nucleosome confers a similar degree of rigidity to histone H4 as is found in the CENP-A-containing nucleosome (SI Fig. 13). Together, these data indicate that substitution into histone H3 of the CATD is sufficient to confer the conformationally more constrained properties that are a hallmark of centromeric CENP-A nucleosomes.
Discussion
A Potential Centromere Mark Generated by the CENP-A-Containing Nucleosome.
Regarding the proposal that CENP-A provides the epigenetic mark to specify the location of the centromere, our finding here that it generates structurally divergent nucleosomes relative to those assembled with H3 provides a means by which this mark is generated. The CENP-A nucleosome is more conformationally constrained within the α2-helix from CENP-A and the α2- and α3-helices from H4, which are predicted to form the interface between CENP-A and histone H4 in three-dimensional space (19). Twelve of the 29 aa that comprise the α2-helix of CENP-A vary from the corresponding positions in canonical H3, and, together with the preceding loop (L1) that varies from the L1 in histone H3 in both primary sequence and length, it forms the CATD. Essential for targeting newly made CENP-A to the centromere, substitution of the CATD into canonical H3 is sufficient to replace the essential function of CENP-A in the maintenance of centromere function (20). Together with earlier evidence that the CATD confers compaction to the prenucleosomal (CENP-A/H4)2 heterotetramer (19), our finding here that it is sufficient to confer structural inflexibility to the nucleosome gives strong support to a model wherein CATD-mediated conformational rigidity provides the cis-acting targeting information for newly made CENP-A as well as the nucleosomal mark that specifies the location for new CENP-A deposition (Fig. 4D). Furthermore, the rigid nucleosome structure generated by the presence of CENP-A occurs in nucleosomes assembled with either centromeric or noncentromeric DNA (Fig. 3), passing the critical test of an epigenetic centromere mark that is known to specify centromere location independent of DNA sequence (2, 3, 13).
Mechanisms for Propagating the Centromere Mark.
The propagation of this potential centromeric mark requires the faithful loading of newly made CENP-A at centromeres during each cell cycle. Because the bulk of new CENP-A deposition does not occur simultaneously with DNA replication (26) and existing CENP-A pools would presumably be equally divided between daughter strands of the replicating DNA, we envision that canonical H3 is deposited in place of CENP-A at intervening sites at which CENP-A has segregated with the opposite strand during S-phase (Fig. 4D). Histone H3 modified by dimethylation at lysine-4 is found interspersed with CENP-A in “stretched chromatin” preparations and in immediate proximity on mitotic chromosomes prepared in the absence of microtubules (27, 28). An attractive extension of this idea is that these particularly modified nucleosomes contain the H3 molecules that are destined to be specifically replaced by newly made CENP-A.
Although the mechanism to eject histone H3 and replace it with newly made CENP-A is unclear, our model of CATD-mediated targeting and marking provides an explanation for how it is faithfully assembled or stabilized at existing centromeres. Potential models for this process include centromere-specific chromatin loading factors (13). Given our findings, the most plausible model is that any CENP-A loading factor would recognize the prenucleosomal (CENP-A/H4)2 heterotetramer via the CATD and deliver it to centromeric chromatin marked by the rigid CENP-A nucleosomes already at adjacent sites. Our data offer experimental support for a pathway leading to CENP-A replenishment that incorporates self-directed, CATD-mediated targeting (Fig. 4D) as the critical step.
The Foundation of Rigid Chromatin Structure at the Inner Kinetochore.
CENP-A nucleosomes are at the foundation of the kinetochore-forming portion of each centromere, organizing into a higher-order disk-shaped chromatin domain (0.5–1 μm in diameter) that forms the attachment site for spindle microtubules (27, 29, 30). We propose that exclusion of H3-containing nucleosomes from the centromeric chromatin domain is driven by the structurally divergent, rigid properties of the CENP-A nucleosome that drive self-association (Fig. 4E). The resulting higher-order chromatin structure composed of coalesced CENP-A-containing nucleosomes is, in turn, a highly rigid structure as observed by monitoring spindle-mediated tension at kinetochores. Although the connection between the spindle and the kinetochore can withstand 700 piconewtons of mechanically generated force (31), and the chromatin between sister kinetochores is stretched >2-fold upon bipolar attachment to the spindle starting in prometaphase (32, 33), CENP-A-containing chromatin remains fixed in size (33).
Such a self-assembly mechanism driving centromere propagation is likely reinforced by recruitment of other constitutive centromere components including the CENP-ANAC (see Fig. 4F) composed of CENP-C, CENP-H, CENP-M, CENP-N, CENP-T, and CENP-U(50) (18). Our finding that the CENP-A-mediated structural differences in centromeric nucleosomes are initiated at the interface between the CATD and H4 in the core of the nucleosome firmly suggests that this initial change propagates a structural divergence from canonical nucleosomes that continues outside this limited domain. Thus, we envision that the CATD likely functions by altering more global physical properties of the nucleosome, as opposed to serving as a direct recognition element, because the amino acid side chains within the conformationally constrained α2-helix portion of the CATD are buried within the center of the nucleosome. In this scenario, one or more CENP-ANAC components would directly recognize and bind to one or a few CENP-A-containing nucleosomes. Future experiments are now required to identify the potential roles of these proteins in specializing centromeric chromatin as a part of the epigenetic mark that is inherited from generation to generation.
Methods
H/D Exchange with Nucleosomes.
Mononucleosomes were reconstituted with all recombinant histones (19, 24) by using the salt gradient dialysis method (22). The DNA sequences and the methods used for their generation and purification are provided in SI Methods. After reconstitution, mononucleosomes were concentrated to 1 mg/ml by using Centricon concentrators (Millipore, Billerica, MA). A total of 15 μl of each nucleosome was mixed with 45 μl of D2O containing 5 mM Tris/50 mM NaCl (pD 7) and incubated for 1 × 102, 1 × 103, 1 × 104, 1 × 105, 2.5 × 105, and 1 × 106 s at 37°C. At these time points, samples were added to vials containing 90 μl of a quench solution (0.8% formic acid/3.2 M guanidine hydrochloride) at 0°C, and samples were immediately frozen at −80°C.
Histone Fragmentation and MS.
Samples were individually melted at 0°C, then injected (145 μl) and pumped through immobilized pepsin columns [250 μl/min, 66-μl column of porcine pepsin coupled to 20AL support (PerSeptive Biosystems, Foster City, CA)]. Protease-generated fragments were collected onto a C18 HPLC column and eluted by a linear acetonitrile gradient (0–50% B in 30 min; 50 μl/min; solvent A, 0.05% TFA; solvent B, 80% acetonitrile/20% water/0.01% TFA), and the effluent was directed to the mass spectrometer with data acquisition in either MS1 profile mode or data-dependent MS2 mode. MS analyses used a LCQ electrospray ion trap type mass spectrometer (ThermoFinnegan, San Jose, CA) operated with capillary temperature at 200°C or an electrospray Micromass Q-Tof mass spectrometer, as previously described (34). The Sequest software program (ThermoFinnegan) was used to identify the likely sequence of the parent peptide ions using nondeuterated samples via tandem MS. Details of analyzing the H/D exchange data are provided in SI Methods.
Supplementary Material
Acknowledgments
We thank J. Kahana, K. Luger, and A. Prunell for gifts of reagents; B. Cottrell, C. Gessner, and S. Li for technical assistance; and L. Jansen, D. Foltz, and P. Maddox for helpful discussions. This research was supported by National Institutes of Health Grants GM29513 and GM074150 (to D.W.C.); National Institutes of Health/National Cancer Institute Innovative Molecular Analysis Technologies Grants CA099835 and CA118595 (to V.L.W.); a postdoctoral fellowship from the American Cancer Society (to B.E.B.); and a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund (to B.E.B.). Salary support for D.W.C. is provided by the Ludwig Institute for Cancer Research. Support for B.E.B. and S.B. is provided by startup funds from the University of Pennsylvania (to B.E.B.).
Abbreviations
- CATD
CENP-A centromere targeting domain
- H/D
hydrogen/deuterium.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700390104/DC1.
References
- 1.Strahl BD, Allis CD. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Cleveland DW, Mao Y, Sullivan KF. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 3.Carroll CW, Straight AF. Trends Cell Biol. 2006;16:70–78. doi: 10.1016/j.tcb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Jiang J, Birchler JA, Parrott WA, Dawe RK. Trends Plant Sci. 2003;8:570–575. doi: 10.1016/j.tplants.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Willard HF, Waye JS. Trends Genet. 1987;3:192–198. [Google Scholar]
- 6.Earnshaw WC, Migeon BR. Chromosoma. 1985;92:290–296. doi: 10.1007/BF00329812. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan BA, Schwartz S. Hum Mol Genet. 1995;4:2189–2197. doi: 10.1093/hmg/4.12.2189. [DOI] [PubMed] [Google Scholar]
- 8.Depinet TW, Zackowski JL, Earnshaw WC, Kaffe S, Sekhon GS, Stallard R, Sullivan BA, Vance GH, Van Dyke DL, Willard HF, et al. Hum Mol Genet. 1997;6:1195–1204. doi: 10.1093/hmg/6.8.1195. [DOI] [PubMed] [Google Scholar]
- 9.du Sart D, Cancilla MR, Earle E, Mao JI, Saffery R, Tainton KM, Kalitsis P, Martyn J, Barry AE, Choo KH. Nat Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. [DOI] [PubMed] [Google Scholar]
- 10.Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, et al. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- 11.Amor DJ, Bentley K, Ryan J, Perry J, Wong L, Slater H, Choo KH. Proc Natl Acad Sci USA. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ventura M, Weigl S, Carbone L, Cardone MF, Misceo D, Teti M, D'Addabbo P, Wandall A, Bjorck E, de Jong PJ, et al. Genome Res. 2004;14:1696–1703. doi: 10.1101/gr.2608804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sullivan BA, Blower MD, Karpen GH. Nat Rev Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- 14.Rajagopalan H, Lengauer C. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 15.Kops GJ, Weaver BA, Cleveland DW. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 16.Sullivan KF, Hechenberger M, Masri K. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T. Proc Natl Acad Sci USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foltz DR, Jansen LET, Black BE, Bailey AO, Yates JR, III, Cleveland DW. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 19.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Cleveland DW. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 20.Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 21.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 22.Luger K, Rechsteiner TJ, Richmond TJ. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 23.Englander SW. J Am Soc Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 25.Vafa O, Sullivan KF. Curr Biol. 1997;7:897–900. doi: 10.1016/s0960-9822(06)00381-2. [DOI] [PubMed] [Google Scholar]
- 26.Shelby RD, Monier K, Sullivan KF. J Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blower MD, Sullivan BA, Karpen GH. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sullivan BA, Karpen GH. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zinkowski RP, Meyne J, Brinkley BR. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rieder CL. Int Rev Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- 31.Nicklas RB. J Cell Biol. 1983;97:542–548. doi: 10.1083/jcb.97.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shelby RD, Hahn KM, Sullivan KF. J Cell Biol. 1996;135:545–557. doi: 10.1083/jcb.135.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maddox P, Straight A, Coughlin P, Mitchison TJ, Salmon ED. J Cell Biol. 2003;162:377–382. doi: 10.1083/jcb.200301088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pantazatos D, Kim JS, Klock HE, Stevens RC, Wilson IA, Lesley SA, Woods VL., Jr Proc Natl Acad Sci USA. 2004;101:751–756. doi: 10.1073/pnas.0307204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.