Abstract
The antioxidant response element (ARE) is a cis-acting regulatory enhancer element found in the 5′ flanking region of many phase II detoxification enzymes. Up-regulation of ARE-dependent target genes is known to have neuroprotective effects; yet, the mechanism of activation is largely unknown. By screening an arrayed collection of ≈15,000 full-length expression cDNAs in the human neuroblastoma cell line IMR-32 with an ARE-luciferase reporter, we have identified several cDNAs not previously associated with ARE activation. A subset of cDNAs, encoding sequestosome 1 (SQSTM1) and dipeptidylpeptidase 3 (DPP3), activated the ARE in primary mouse-derived cortical neurons. Overexpression of SQSTM1 and DPP3 in IMR-32 cells stimulated NF-E2-related factor 2 (NRF2) nuclear translocation and led to increased levels of NAD(P)H:quinone oxidoreductase 1, a protein which is transcriptionally regulated by the ARE. When transfected into IMR-32 neuroblastoma cells that were depleted of transcription factor NRF2 by RNA interference, SQSTM1 and DPP3 were unable to activate the ARE or induce NAD(P)H:quinone oxidoreductase 1 expression, indicating that the ARE activation upon ectopic expression of these cDNAs is mediated by NRF2. Studies with pharmacological inhibitors indicated that 1-phosphatidylinositol 3-kinase and protein kinase C signaling are essential for activity. Overexpression of these cDNAs conferred partial resistance to hydrogen peroxide or rotenone-induced toxicity, consistent with the induction of antioxidant and phase II detoxification enzymes, which can protect from oxidative stress. This work and other such studies may provide mechanisms for activating the ARE in the absence of general oxidative stress and a yet-unexploited therapeutic approach to degenerative diseases and aging.
Keywords: genome-wide screen, oxidative stress, neuroprotection
Oxidative stress is implicated in the pathogenesis of many age-related diseases, including neurodegenerative disorders, such as Alzheimer's and Parkinson's disease, and aging itself (1). In humans, the antioxidant response element (ARE) regulates the expression of a number of cytoprotective antioxidant enzymes and scavengers, which contribute to the endogenous defense against oxidative stress. The ARE is a cis-acting regulatory enhancer element (core sequence: 5′-GTGACnnnGC-3′) found in the 5′ flanking region of many phase II detoxification enzymes and is activated by reactive oxygen species, as well as other electrophilic agents (2). Genes regulated by the ARE include heme oxygenase-1, GSTs, and NAD(P)H:quinone oxidoreductase 1 (NQO1). It has been shown that activation of the ARE protects neuroblastoma cells, astrocytes, and neurons from oxidative damage (3–5).
The molecular mechanism of ARE activation is largely unknown. However, the central transcription factor involved in the induction of phase II enzymes has been shown to be NF-E2-related factor 2 (Nrf2). Nrf2 knockout mice show reduced expression of glutathione biosynthetic genes (6) and GSTs (7), diminished detoxification capabilities (8), decreased responsiveness to chemoprotective agents (9), and enhanced susceptibility to oxidative stress-induced cell death (10–12). Conversely, Nrf2 overexpression in vitro and in vivo protects from oxidative stress (10, 11). Nrf2 is bound to the cytoplasmic repressor protein Keap1 and, upon activation, translocates into the nucleus and transcriptionally activates ARE-dependent genes after recruiting Maf proteins (2). The upstream regulatory mechanisms by which ARE-activating signals are linked to Nrf2 remain to be fully elucidated. It has been demonstrated that reactive sulfhydryl groups of Keap1 are sensors for induction of phase II genes (13), leading to the proposal that the Nrf2/Keap1 interaction represents a cytoplasmic sensor for oxidative stress. However, 1-phosphatidylinositol 3-kinase (PI3K), MAPKs, and protein kinase C (PKC) have also been implicated in ARE activation (14–17), suggesting that multiple signaling mechanisms may be involved. A deeper understanding of the molecular mechanisms governing the response to oxidative stress is necessary to exploit this signaling pathway therapeutically. Consequently, we have carried out a genome-wide, high-throughput screen to identify previously unrecognized ARE activators. Genome-scale screens of this type with spatially arrayed cDNA and siRNA libraries have proven powerful for the functional analysis of various pathways involved in inflammation, cancer, insulin signaling, and other biological processes (18).
Results
High-Throughput Screen of Arrayed cDNA Library for ARE Activators.
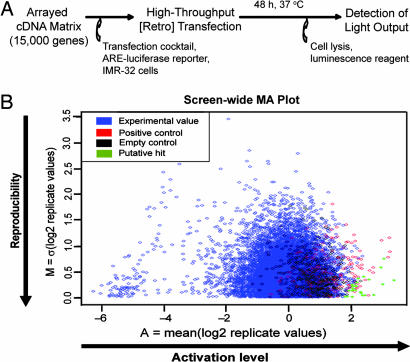
A cDNA library consisting of ≈15,000 full-length human expression cDNAs arrayed in 384-well plates was screened for activators of the ARE (19). The cDNA collection, covering approximately half of the human genome, was obtained from OriGene Technologies (Rockville, MD); each cDNA was cloned downstream of a CMV promoter. The library was transfected into IMR-32 human neuroblastoma cells along with a human NQO1-ARE reporter construct consisting of the luciferase gene under the control of the ARE-containing promoter, as described in ref. 20. The IMR-32 cell line has been used as a cellular model of oxidative stress (3, 14, 20). After 48 h, luminescence was detected in each well as a measure of ARE activity (Fig. 1A). The screen was carried out in duplicate to evaluate the reproducibility of the results, and a constitutively active PI3K construct (21), which is known to activate the ARE in IMR-32 cells (14), was used as a positive control (five-fold average activation over background). cDNAs that showed high activation and high reproducibility were selected for further analysis (Fig. 1B, lower right corner, green).
Fig. 1.
Genome-wide cDNA overexpression screen for ARE activators. (A) General high-throughput screening procedure. Approximately 15,000 expression cDNAs, normalized and arrayed in 384-well plates, were transfected into IMR-32 human neuroblastoma cells along with an ARE–luciferase reporter construct. After 48-h incubation, luciferase activity was assessed by measuring luminescence output per well. (B) Screen-wide MA plot. The screen was carried out in duplicate, and M (a measure of screen-to-screen variation; σ = standard deviation) was plotted as a function of A (a measure of the mean ARE activation from both screens). The cDNAs that strongly activated the ARE in a reproducible manner were investigated further (indicated in green, lower right corner).
Proteins encoded by eight cDNAs reproducibly activated the ARE by 5- to 46-fold over background. These cDNAs encode sequestosome 1 (SQSTM1), D-site of albumin promoter binding protein (DBP), dipeptidylpeptidase 3 (DPP3), BCL2-like 1 (BCL2L1; longer isoform, Bcl-xL), kinesin family member 26B (KIF26B), cAMP-responsive element binding protein-regulated transcription coactivator 1 (TORC1), myeloid cell leukemia sequence 1 (MCL1; longer isoform, Mcl-1l), and splicing factor, arginine/serine-rich 10 (SFRS10). The ubiquitin-binding protein SQSTM1 and the antiapoptotic isoforms of the BCL2-related proteins BCL2L1 (Bcl-xL) and MCL1 represent previously characterized prosurvival gene products. SQSTM1 is induced by oxidative stress and mediates diverse signaling pathways associated with cell stress, survival, and inflammation (22–24). It colocalizes with protein aggregates found in Parkinson's, Pick's, and Alzheimer's disease (25–27), and was recently reported to prevent huntingtin-induced cell death (28). Bcl-xL regulates the outer mitochondrial membrane channel opening and consequently controls the production of reactive oxygen species and cytochrome c release by mitochondria (29). Overexpression of Bcl-xL has been shown to inhibit 6-hydroxydopamine-induced death in a Parkinson's disease-like model in a neuroblastoma cell line (30). The longer isoform of MCL1 is a viability-promoting member of the BCL2 family that reduces cell damage-induced release of mitochondrial cytochrome c. It exerts its antiapoptotic properties by inhibiting mitochondrial Ca2+ signals (31). Two other cDNAs encode transcription-associated factors (DBP and TORC1). DBP is a basic leucine zipper transcription factor (32); TORC1 (MECT1) is a cAMP-responsive element binding protein-dependent transcriptional coactivator. The fusion product of TORC1 with MAML2 has transforming, cAMP-responsive element binding protein-dependent activity (33) and is required for the growth of mucoepidermoid salivary gland tumors (34). DPP3 is a zinc metallo-exopeptidase with broad substrate specificity implicated in various disease processes, such as cancer and inflammation (35–37). One cDNA, SFRS10, encodes a splicing factor involved in exon 10 splicing of tau (38, 39), a microtubule-stabilizing protein that is hyperphosphorylated and forms neurofibrillary tangles in Alzheimer's disease. KIF26B contains a kinesin motor domain that is suggestive of a microtubule-dependent molecular motor function. This gene product has been postulated to play a role in embryogenesis because of its preferential expression in the embryo (40).
Confirmation of ARE Activation by the cDNAs.
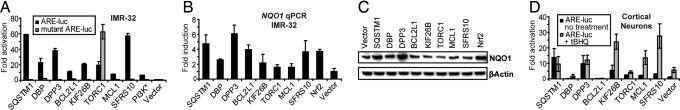
ARE activators that were selected after the primary screen were retested in 24-well plate format, and activities were normalized for transfection efficiency. The cDNAs activated the reporter 7- to 59-fold over the vector control, which compares favorably to the positive control, constitutively active PI3K (PI3K*), which activated 7-fold (Fig. 2A). SQSTM1, SFRS10, and DPP3 were the strongest activators but still below the ARE activation induced by Nrf2 overexpression (>200-fold) (41). The confirmed ARE activators were also transfected with a mutant ARE reporter construct (20, 41) to test for ARE sequence specificity (Fig. 2A). This reporter contains a GC→AT mutation in the ARE core sequence known to abolish Nrf2-mediated ARE activation (20, 41). Only one of the eight cDNAs (TORC1) activated the ARE nonspecifically. Finally, all cDNAs activated the wild-type ARE reporter construct to the same extent in the presence of excess antioxidant (1 mM N-acetyl cysteine), indicating that the cDNAs do not activate the ARE by inducing oxidative stress [supporting information (SI) Table 1].
Fig. 2.
Confirmation of putative screening hits in IMR-32 cells and mouse primary cortical culture. (A) Transcriptional ARE activation in IMR-32 cells with wild-type and mutant enhancer elements. IMR-32 cells were cotransfected with cDNAs, the ARE-luciferase reporter (wild-type or mutant), and actin-lacZ for normalization in 24-well plates. Luminescence was detected 48 h later; normalized values are given (n = 6). Eight cDNAs showed equal or higher activity than the positive control, PI3K*. (B) Effect of cDNA overexpression in IMR-32 cells on NQO1 transcript levels as analyzed by quantitative real-time PCR (qPCR). The cDNAs were introduced by lipofection, and then total RNA was isolated 48 h later, reverse-transcribed to cDNA, and subjected to TaqMan analysis (n = 3). GAPDH expression was used as internal control for normalization. Overexpression of several cDNAs increased NQO1 levels to the same extent as Nrf2 overexpression. (C) Induction of NQO1 upon cDNA overexpression in IMR-32 cells as analyzed by Western blot analysis. cDNAs were transfected by using lipofection, and proteins were isolated 48 h later, resolved by SDS/PAGE, and subjected to Western blot analysis for NQO1. SQSTM1 and DPP3 induced NQO1 most strongly and to a comparable extent as Nrf2. A representative blot (n = 4) is shown. (D) Transcriptional ARE activation in mouse primary cortical culture. After 5 days, cells were transfected with the reporter mixture (ARE–luciferase, CMV-lacZ, and cDNA), and 24 h later cells were treated either with 10 μM tBHQ or with vehicle control. After an additional 24 h, luminescence was detected. Normalized values are given (n = 3). Only a subset of cDNAs with activity in IMR-32 cells showed activity in primary cells. The ARE activity of SQSTM1 and DPP3 could not be further augmented by tBHQ.
Induction of Expression of Antioxidant Enzymes.
Next, it was determined whether cDNA overexpression also induces the expression of endogenous ARE-regulated genes. Quantitative PCR (qPCR) after reverse transcription (RT) was used to determine transcript levels of NQO1, which is transcriptionally regulated by ARE (14). NQO1 prevents the reduction of quinones, which leads to the production of radical species and has been linked with Alzheimer's disease (42). cDNAs were transfected into IMR-32 cells (≈70% transfection efficiency), and RNA was extracted 48 h later and subjected to RT-qPCR analysis of NQO1 mRNA levels. Overexpression of most cDNAs significantly increased NQO1 expression; several cDNAs (SQSTM1, DPP3, BCL2L1, and SFRS10; 3.7- to 6.1-fold) had activities comparable to Nrf2 (3.7-fold) under the conditions used (Fig. 2B). For comparison, the well-characterized ARE activator tert-butylhydroquinone (tBHQ) (10 μM, 24-h treatment) appeared to be a stronger inducer of NQO1 expression with a 13.0-fold increase over the DMSO control, which is similar to the reported value (3, 43) (however, this is at least partly due to the fact that all cells are affected by small molecule treatment in contrast to cDNA overexpression, which suffers from higher background arising from nonquantitative transfection efficiency). Western blot analysis was carried out to determine whether the effects on gene expression correlate with the levels of ARE-regulated enzymes such as NQO1. cDNAs were transiently transfected into IMR-32 cells, proteins were extracted 48 h later, and NQO1 expression was assayed by Western blot analysis. SQSTM1 and DPP3 overexpression caused a 2.6- and 3.6-fold increase in NQO1 protein levels, respectively, which is similar to that obtained for Nrf2 overexpression (2.3-fold, Fig. 2C). Overexpression of all other cDNAs resulted in negligible or no apparent increases of NQO1 enzyme levels (Fig. 2C), indicating a nonlinear relationship of transcript and protein levels. These results led us to focus on SQSTM1 and DPP3 activity. To further characterize the genes up-regulated by SQSTM1 and DPP3, genome-wide gene expression analysis was carried out. At the two-fold level, overexpression of each cDNA up-regulated only the expected NQO1, the gene encoding chaperone CCT5, and two noncoding transcripts: microRNA 21 and noncoding RNA MALAT1 (SI Table 2). A number of other detoxification and antioxidant stress genes, including aldo-keto reductases (AKR1C1 and AKR1C2), a thioredoxin (TXNDC11), and ferritin (FTH1), were up-regulated 1.4- to 2.0-fold upon overexpression of SQSTM1 and DPP3, some of which were verified by RT-qPCR (SI Table 2).
ARE Activation in Primary Cortical Culture.
Next, the activity of the cDNAs was tested in a more relevant cellular context, specifically mouse primary cortical neurons. The cDNAs and ARE–luciferase construct were cotransfected by lipofection and ARE activity was assayed 48 h later by recording luminescence. ARE activation was normalized on the basis of β-galactosidase activity arising from additional cotransfection with CMV-lacZ. A subset of cDNAs (SQSTM1, DPP3, KIF26B, TORC1, MCL1, and SFRS10) activated the ARE reporter 2- to 14-fold relative to the vector control (Fig. 2D). In the case of SQSTM1 and DPP3, activation could not be further increased by the addition of tBHQ, which functions by inducing Nrf2 translocation (41). In contrast, KIF26B, MCL1, and SFRS10 activated the ARE in a synergistic fashion with tBHQ, suggesting different modes of action. The ARE activity of DBP and BCL2L1 could not be confirmed in primary cells, suggesting that their physiological role may not be relevant to ARE activation.
Nuclear Translocation of NRF2 Is Promoted upon Overexpression of SQSTM1 or DPP3.
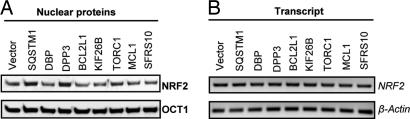
Because ARE activation occurs through the transcription factor NRF2, which translocates into the nucleus to induce target gene expression, the subcellular localization of NRF2 was determined after cDNA transfection. Nuclear extracts were prepared by standard methods (44), and NRF2 levels were measured by immunoblot analysis. Consistent with the induction of NQO1, an approximately two-fold enrichment of NRF2 was found in the nucleus 48 h after transfection with SQSTM1 and DPP3 encoding cDNAs (Fig. 3A). Overexpression of the other cDNAs (DBP, BCL2L1, KIF26B, TORC1, MCL1, and SFRS10) did not significantly affect nuclear NRF2 content (Fig. 3A), which is suggestive of alternative modes of action. Moreover, comparison of NRF2 transcript levels in whole-cell lysates by semiquantitative RT-PCR analysis did not show a significant difference among cells transfected with cDNAs or vector control (Fig. 3B), suggesting that the major route of ARE activation by SQSTM1 and DPP3 is through promotion of NRF2 translocation. Apparent total NRF2 protein levels increased slightly upon SQSTM1 and DPP3 overexpression (SI Fig. 6), which likely reflects the strong enrichment of NRF2 in the nucleus where it is more stable and not susceptible to proteasomal degradation as in the cytoplasm. SQSTM1 and DPP3 may additionally stabilize NRF2 as reported for tBHQ (45).
Fig. 3.
Analysis of NRF2 translocation upon cDNA overexpression in IMR-32 cells. (A) Enrichment of NRF2 in the nucleus by SQSTM1 and DPP3 overexpression as determined by immunoblot analysis. Nuclear extracts prepared with the NE-PER reagent kit (Pierce) 48 h after transfection were resolved by SDS/PAGE, and the Western blot was probed with NRF2 antibody. A representative blot (n = 3) is shown. SQSTM1 and DPP3 significantly increased NRF2 content in the nucleus. Levels of the nuclear protein OCT1 did not change. (B) Effect of cDNA overexpression on NRF2 transcript levels. Total RNA from IMR-32 cells was harvested 48 h after cDNA transfection. Semiquantitative RT-PCR suggested that NRF2 transcript levels did not change significantly. Data shown are representative of multiple experiments (n = 3) in the linear amplification range.
NRF2 siRNA Attenuates ARE Activity and NQO1 Expression.
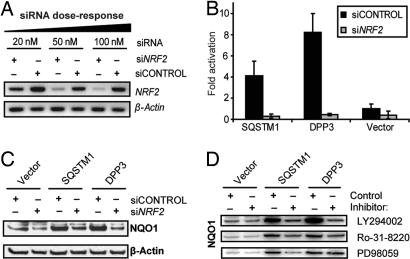
To ascertain that NRF2 is the major transcription factor responsible for ARE activation and NQO1 induction upon cDNA overexpression, four pooled siRNAs designed by Dharmacon (Lafayette, CO) (46) were used to selectively knock down NRF2 transcript levels. Dose-response analysis indicated that a 50 nM concentration of specific siRNAs after 48 h reproducibly decreased NRF2 transcript levels in IMR-32 cells by >70%, as determined by semiquantitative RT-PCR followed by densitometry (Fig. 4A), and also led to a reduction in NRF2 protein levels after 48 h by >80% on the basis of immunoblot analysis (SI Fig. 6). IMR-32 cells were then depleted of NRF2 by transfecting the validated siRNAs (50 nM), and cDNAs encoding SQSTM1 or DPP3 were cotransfected along with CMV–GFP to monitor DNA transfection efficiency (≈70% for all cDNAs). Cotransfecting the ARE–luciferase construct enabled the measurement of residual transcriptional ARE activity in NRF2-depleted cells. ARE activity was completely abolished after 48 h in the presence of either SQSTM1 or DPP3 and NRF2-specific siRNAs (Fig. 4B). In addition, protein lysates were collected 48 h after transfection, and immunoblot analysis for NQO1 was performed. The expression of NQO1 was completely abrogated in NRF2-depleted cells transfected with SQSTM1 or DPP3 but not in cells cotransfected with nontargeting control siRNAs (Fig. 4C). This result indicated that the phase II enzyme expression induced by SQSTM1 and DPP3 is mediated by transcription factor NRF2.
Fig. 4.
Effect of siRNAs targeting NRF2 and effects of kinase inhibitors in IMR-32 cells. (A) Dose-response analysis for siRNAs by using RT-PCR. SiLentFect-mediated transfection of 50 nM siRNAs almost completely abrogated NRF2 transcript levels after 48 h in a reproducible manner (n = 4). (B) Effect of SQSTM1 and DPP3 overexpression in NRF2-depleted IMR-32 cells. The cDNAs encoding SQSTM1 and DPP3 (1.4 μg) were cotransfected with ARE-luciferase reporter (1.0 μg) and with siRNAs against NRF2 or control siRNAs (50 nM) in six-well plate format. After 48 h, luciferase activity was measured. DNA transfection efficiency was monitored by cotransfecting CMV–GFP (0.2 μg) and was similar in each well. SQSTM1 and DPP3 were unable to activate the ARE in cells transfected with siRNAs against NRF2 (n = 4). Note that ARE activation by SQSTM1 and DPP3 in the control cells is lower than that shown in Fig. 2A because less cDNA was transfected in this experiment, consistent with a dose-dependency of ARE activation. (C) Analysis of SQSTM1 and DPP3 mediated induction of NQO1 in NRF2-depleted IMR-32 cells. The cDNAs encoding SQSTM1 and DPP3 (1.4 μg) were cotransfected with siRNAs against NRF2 or control siRNAs (50 nM) in six-well plate format. Equal DNA transfection efficiency was obtained by monitoring GFP expression upon cotransfection with CMV–GFP (0.2 μg). SQSTM1 and DPP3 were unable to induce NQO1 expression in cells cotransfected with siRNAs targeting the NRF2 transcript. Nontargeting siRNAs did not diminish the ability of SQSTM1 and DPP3 to induce NQO1. Control protein levels (β-actin) did not change across experimental conditions. A representative blot (n = 3) is shown. (D) Effect of pharmacological kinase inhibitors on SQSTM1- and DPP3-induced NQO1 expression. IMR-32 cells were seeded in six-well plates (600,000 cells per well) and 1 day later were transfected with cDNA (1.6 μg) and CMV–GFP (0.4 μg) by using siLentFect. After 24 h, cells were treated for additional 24 h with PI3K inhibitor LY294002 (25 μM), PKC inhibitor Ro-31-8220 (1 μM), and MEK1 inhibitor PD98059 (50 μM). Concentrations used have been shown to be effective (14, 17). Representative blots (n = 3) are shown.
Effect of Pharmacological Inhibitors.
To reveal important upstream factors governing the activation of the Nrf2–ARE pathway upon SQSTM1 or DPP3 overexpression, we examined kinase signaling implicated in ARE activation. In particular, ARE activity of tBHQ in IMR-32 cells is known to be PI3K-dependent and therefore attenuated by PI3K inhibitors (14). Consequently, the effect of a selective pharmacological inhibitor of PI3K, LY294002, on the ability of SQSTM1 and DPP3 to induce the expression of endogenous NQO1 protein was determined with an established effective concentration (25 μM) (14). Both cDNAs were unable to induce NQO1 in the presence of LY294002 (Fig. 4D). A similar result was obtained with a selective PKC inhibitor (Ro-31-8220) at a concentration of 1 μM (17). However, inhibition of MAPK signaling with the MEK1 inhibitor PD98059 (50 μM) only attenuated NQO1 expression induced by SQSTM1 but not by DPP3 (Fig. 4D). For comparison, PD98059 (50 μM) is known to have no effect on tBHQ-induced ARE activation in this cell type, although clearly inhibiting phosphorylation of ERK1/2 at this concentration (14).
Protection from Oxidative Stress.
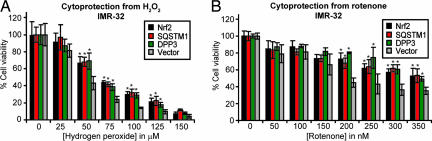
ARE activation and subsequent induction of antioxidant factors is expected to confer resistance to oxidative stress. To test whether overexpression of SQSTM1 and DPP3 has the ability to protect from toxicity induced by oxidative insult, IMR-32 cells were treated 48 h after cDNA transfection with hydrogen peroxide for 6 h or the mitochondrial complex I inhibitor rotenone (10) for 12 h, and cell viability was measured. Overexpression of both SQSTM1 and DPP3 attenuated the cytotoxicity to a similar extent as Nrf2 overexpression and approximately doubled the cell viability after treatment with 100 μM hydrogen peroxide (Fig. 5A) or 200–300 nM rotenone (Fig. 5B). The magnitude of cytoprotection from hydrogen peroxide by Nrf2 overexpression closely matches reports for Nrf2 overexpression in the immortalized mouse hippocampal cell line HT-22 (47).
Fig. 5.
Nrf2, SQSTM1 and DPP3 mediate protection from oxidative stress in vitro. In 24-well plate format with siLentFect, 180,000 IMR-32 cells were transfected with corresponding cDNAs and with 0.5 μg of vector control. After 48 h, cells were treated with various concentrations of hydrogen peroxide for 6 h (A) or rotenone for 12 h (B) (n = 4). Cell viability was assessed by using the CellTiter-Glo assay kit (Promega). Overexpression of Nrf2, SQSTM1, or DPP3 attenuated the toxic effects of both stressors. ∗ indicates statistical significance compared with vector (P < 0.05).
Discussion
The activation of the ARE in the absence of general oxidative stress could provide a yet-unexploited therapeutic approach for the treatment of various neurodegenerative diseases, stroke, and aging. To this end, a genome-wide overexpression screen was carried out in IMR-32 cells to identify previously unrecognized mediators of ARE activity. A reporter gene-based genomic approach yielded several gene products hitherto unknown to activate the ARE. Two of the identified cDNAs (encoding DBP and BCL2L1) were unable to elicit a transcriptional response in primary neuronal culture, indicating cell-type specificity, whereas TORC1 is regarded as an overexpression artifact because it activated both ARE and a mutant reporter in a nondiscriminatory fashion. For several cDNAs, the strong activation evident in the reporter gene assay in IMR-32 cells did not translate into significant up-regulation of the downstream gene NQO1 or noticeable induction of NRF2 nuclear translocation. Most importantly, two of the strongest ARE activators in the reporter gene assay in IMR-32 neuroblastoma cells, viz. SQSTM1 and DPP3, were validated in a variety of secondary assays and emerged as prime candidates for further study.
Upon ectopic expression, both SQSTM1 and DPP3 potently activated the ARE in a reporter gene assay in primary cortical neuronal culture, suggesting a physiological relevance of these gene products in ARE activation. Consistent with the transcriptional activation of the ARE, overexpression of SQSTM1 and DPP3 in IMR-32 cells also increased the expression levels of NQO1, a phase II detoxification enzyme regulated by the ARE. Overexpression of both SQSTM1 and DPP3 caused NRF2 to translocate into the nucleus, suggesting that the transcription factor NRF2 mediates the ARE activation induced by these two proteins. Consistent with this notion, siRNAs targeting NRF2 completely suppressed their ability to activate the ARE and consequently induce NQO1 expression. Thus, both gene products are previously unrecognized regulators in the canonical Nrf2–ARE pathway. Experiments with pharmacological inhibitors indicated that the upstream mechanism is controlled by PI3K and PKC kinase-signaling pathways, in agreement with previous studies that show that these pathways play a role in ARE activation (15–17). And finally, overexpression of the NRF2-dependent ARE activators SQSTM1 and DPP3 protects neuroblastoma cells from oxidative stress, demonstrating that these gene products play a functional role in the antioxidant response.
SQSTM1 is a known stress-response protein that is up-regulated upon a variety of stress stimuli. Although SQSTM1 is known to be induced by ARE activation (22, 48), it was not known that SQSTM1 in turn can act as a positive regulator of the ARE. The accumulation of SQSTM1 has been associated with neurodegenerative diseases because it is a component of toxic aggregates in the brains of those with Parkinson's, Alzheimer's, and Pick's disease (26, 27). Recent evidence suggests that SQSTM1 has protective properties (28). One mechanism whereby SQSTM1 may protect neuronal cells from toxicity of misfolded proteins is by enhancing aggregate formation (25). Our results support a cytoprotective role of SQSTM1 and suggest that the beneficial, prosurvival effects of SQSTM1 may partially be mediated through activation of the ARE. PI3K likely transmits this effect because selective PI3K inhibitors have been shown to inhibit cell survival induced by SQSTM1 (49), and we have demonstrated that SQSTM1-induced NQO1 expression is abrogated by the PI3K inhibitor LY294002. PKC and MAPK signaling also appear to be important mediators of NQO1 induction by SQSTM1. The PKC dependence may be explained by the ability of SQSTM1 to directly bind to atypical PKC through its acidic interaction domain (23).
DPP3 is a cytoplasmic serine protease with broad specificity (35) that has been associated with various diseases. Its presence in neutrophils suggests that it plays a physiological role in inflammation (37). It also appears to be one of the most important enkephalin-degrading enzymes in the central nervous systems and thus a regulator of pain (37). Furthermore, DPP3 displays high activity for angiotensin II (50), which plays a role in hypertension. In addition, DPP3 activity is increased in malignant ovarian carcinomas and correlates with aggressiveness of the tumor (36). Here we show that DPP3 also exhibits ARE-activating and cytoprotective properties, which may be related to some of the known activities of DPP3 mentioned above. For example, the Nrf2–ARE pathway has been linked to inflammation (51), whereas its antiapoptotic properties can be tied to cancer progression. Our results implicate the PI3K and PKC signaling pathways in DPP3-mediated ARE activation and exclude a role of MAPK signaling in IMR-32 cells. This result parallels reported findings for tBHQ (14) and differs from our results obtained for SQSTM1. Consequently, it is likely that several kinase-signaling pathways that converge upstream of Nrf2 regulate the ARE activity exerted by SQSTM1 and DPP3.
In summary, we have identified two proteins, SQSTM1 and DPP3, which activate the ARE by inducing nuclear translocation of NRF2, a transcription factor regarded as “multiorgan protector” because of its array of cytoprotective target genes in various organs (52). The ARE activity is likely responsible for the neuroprotective properties upon overexpression in cell culture. Although clearly sufficient for ARE activation, both SQSTM1 and DPP3 do not appear to be necessary components in the oxidative stress-response pathway in IMR-32 cells. Preliminary data indicate that tBHQ can still induce NQO1 expression in cells that are depleted of SQSTM1 or DPP3 by RNA interference. In contrast, this ability was almost completely abrogated in NRF2-depleted cells (SI Fig. 7).
Materials and Methods
Plasmids, Cell Culture, and Assay Conditions.
Expression cDNAs in vector pCMV-XL were obtained from OriGene. Wild-type and mutant ARE–luciferase constructs have been described in refs. 20 and 41. For sequence information, see SI Text. IMR-32 cells (ATCC) were maintained at 37°C humidified air (5% CO2) and assayed in high glucose DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% FBS (HyClone, Logan, UT). Cell culture and assay conditions for mouse primary cortical culture have been described in ref. 53.
Genome-Wide Overexpression Screen.
A library of ≈15,000 expression cDNAs individually arrayed in 384-well plates (62.5 ng per well) was transfected into IMR-32 cells by using FuGENE6 (Roche, Indianapolis, IN) along with the ARE–luciferase construct with high-throughput retro-transfection and luminescence analyzed 48 h later (see SI Text). Relative light intensities were plate-normalized, and signal averages were determined for duplicate screens.
Confirmation and Specificity Studies.
For confirmation studies in 24-well plates, cDNAs (1.56 μg) were transfected into IMR-32 cells (200,000 cells) by using FuGENE6 along with actin-lacZ (for normalization, 200 ng) and either the wild-type or mutant ARE–luciferase construct (625 ng). Luciferase and β-galactosidase activities were measured after 48 h by using Bright-Glo (Promega, Madison, WI) and the Gal-Screen System (Applied Biosystems, Foster City, CA), respectively, and normalized ARE activities were expressed as a ratio of both activities.
Mouse Primary Cortical Cultures.
Primary neuronal cultures were prepared as described in ref. 53 from E19 embryos. On day 5 in vitro, neurons were transfected with a mixture of DNA (150 ng of cDNA, 50 ng of ARE–luciferase reporter, and 50 ng of CMV-lacZ per well) by using FuGENE6. On the day after transfection, cells were treated with vehicle or 10 μM tBHQ. After an additional 24-h incubation, cells were harvested, and luciferase and β-galactosidase activities were recorded as described in ref. 41. Data are shown as the ratio of luciferase to β-galactosidase activities.
Immunoblot Analysis.
Transfections were carried out with siLentFect (Bio-Rad, Hercules, CA) (see SI Text). Forty-eight hours after transfection, whole-cell lysates were prepared by using PhosphoSafe lysis buffer (Novagen, Madison, WI). Nuclear and cytoplasmic proteins were separated by using the NE-PER reagent kit (Pierce, Rockford, IL). Lysates containing equal amounts of protein were separated by SDS/PAGE, transferred to PVDF membranes, probed with antibodies, and detected with the ECL (Amersham, Piscataway, NJ) or Supersignal Femto Western blotting kit (Pierce). Nrf2, Oct1, and secondary anti-goat antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA), NQO1 antibody was obtained from Abcam (Cambridge, MA), and β-actin, secondary anti-rabbit and anti-mouse antibodies were obtained from Cell Signaling (Beverly, MA).
RNA Extraction and Semiquantitative RT-PCR.
Transfections were carried out with siLentFect (Bio-Rad) (see SI Text). Total RNA was extracted 48 h later by using the RNeasy mini kit (Qiagen, Valencia, CA). cDNA was synthesized by using SuperScript II reverse transcriptase (Invitrogen) and Oligo(dT)12–18 primer (Invitrogen). For semiquantitative PCR, amplification was carried out with Platinum PCR SuperMix High Fidelity (Invitrogen). Primer sequences were as follows: β-actin (forward, 5′-AGAGCTACGAGCTGCCTGAC-3′; reverse, 5′-AGTACTTGCGCTCAGGAGGA-3′), NRF2 (forward, 5′-GGCCCATTGATGTTTCTGAT-3′; reverse, 5′-AGCGGCTTGAATGTTTGTCT-3′).
GeneChip Analysis and RT-qPCR.
Duplicate sample preparation for GeneChip analysis was carried out according to the protocol detailed by Affymetrix (Santa Clara, CA). For qPCR, RNA was reverse transcribed to cDNA by using the same method as described for semiquantitative RT-PCR. Real-time PCR with TaqMan chemistry was performed according to the manufacturer's instructions (Applied Biosystems, Foster City, CA) in triplicate analysis. For details, see SI Text.
RNA Interference Experiments.
Nontargeting control siRNA and siGENOME SMARTpool siRNA reagents were obtained from Dharmacon. Forty-eight hours after siLentFect-mediated transfection, RNA or proteins were harvested by using the RNeasy kit (Qiagen) or PhosphoSafe lysis buffer (Novagen), respectively, and subjected to RT-PCR or immunoblot analysis.
Inhibitor Studies.
Twenty-four hours after transfection with cDNAs, IMR-32 cells were treated with LY294002 (25 μM), PD98059 (50 μM), or Ro-31-8220 (1 μM). After 24 h of treatment, proteins were harvested and subjected to immunoblot analysis.
Protection Assays.
IMR-32 cells were batch-transfected with cDNA (0.5 μg per well) and CMV–GFP (0.1 μg per well) by using siLentFect and seeded into 24-well plates (180,000 cells per well). After 48 h of incubation, cells were treated with various doses of hydrogen peroxide for 6 h or rotenone for 12 h. Subsequently, cell viability was measured by using CellTiter-Glo (Promega, Madison, WI).
Detailed experimental procedures are provided in SI Text.
Supplementary Material
Acknowledgments
We thank Sumit Chanda and Tony Orth for providing the cDNA library and screening infrastructure; Phillip McClurg for statistical analysis; and Stephen Ho and Paul DeJesus for technical assistance (all at the Genomics Institute of the Novartis Research Foundation). Funding was provided by the Evelyn F. McKnight Brain Research Grant Program (to H.L.), the American Heart Association (to H.L.), the Novartis Research Foundation (to P.G.S.), the Ellison Medical Foundation (to P.G.S.), and National Institute on Environmental Health Sciences Grants ES10042 (to J.A.J.) and ES08089 (to J.A.J.).
Abbreviations
- ARE
antioxidant response element
- NQO1
NAD(P)H:quinone oxidoreductase 1
- Nrf2
NF-E2-related factor 2
- NRF2
human NF-E2-related factor
- PI3K
1-phosphatidylinositol 3-kinase
- qPCR
quantitative PCR
- RT
reverse transcription
- SQSTM1
sequestosome 1
- tBHQ
tert-butylhydroquinone.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data reported in this paper has been deposited in the Gene Expression Omnibus database (accession no. GSE6451).
This article contains supporting information online at www.pnas.org/cgi/content/full/0700898104/DC1.
References
- 1.van Muiswinkel FL, Kuiperij HB. Curr Drug Targets CNS Neurol Disord. 2005;4:267–281. doi: 10.2174/1568007054038238. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen T, Sherratt PJ, Pickett CB. Annu Rev Pharmacol Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Lee JM, Johnson JA. J Biol Chem. 2002;277:388–394. doi: 10.1074/jbc.M109380200. [DOI] [PubMed] [Google Scholar]
- 4.Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. J Biol Chem. 2003;278:12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 5.Johnson DA, Andrews GK, Xu W, Johnson JA. J Neurochem. 2002;81:1233–1241. doi: 10.1046/j.1471-4159.2002.00913.x. [DOI] [PubMed] [Google Scholar]
- 6.McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, Wolf CR, Cavin C, Hayes JD. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- 7.Chanas SA, Jiang Q, McMahon M, McWalter GK, McLellan LI, Elcombe CR, Henderson CJ, Wolf CR, Moffat GJ, Itoh K, Yamamoto M, Hayes JD. Biochem J. 2002;365:405–416. doi: 10.1042/BJ20020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan K, Han XD, Kan YW. Proc Natl Acad Sci USA. 2001;98:4611–4616. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, Kensler TW. Proc Natl Acad Sci USA. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JM, Shih AY, Murphy TH, Johnson JA. J Biol Chem. 2003;278:37948–37956. doi: 10.1074/jbc.M305204200. [DOI] [PubMed] [Google Scholar]
- 11.Calkins MJ, Jakel RJ, Johnson DA, Chan K, Kan YW, Johnson JA. Proc Natl Acad Sci USA. 2005;102:244–249. doi: 10.1073/pnas.0408487101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burton NC, Kensler TW, Guilarte TR. Neurotoxicology. 2006;27:1094–1100. doi: 10.1016/j.neuro.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 13.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang MI, Kobayashi A, Yamamoto M, Kensler TW, Talalay P. Proc Natl Acad Sci USA. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JM, Hanson JM, Chu WA, Johnson JA. J Biol Chem. 2001;276:20011–20016. doi: 10.1074/jbc.M100734200. [DOI] [PubMed] [Google Scholar]
- 15.Yu R, Lei W, Mandlekar S, Weber MJ, Der CJ, Wu J, Kong AN. J Biol Chem. 1999;274:27545–27552. doi: 10.1074/jbc.274.39.27545. [DOI] [PubMed] [Google Scholar]
- 16.Huang HC, Nguyen T, Pickett CB. J Biol Chem. 2002;277:42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 17.Numazawa S, Ishikawa M, Yoshida A, Tanaka S, Yoshida T. Am J Physiol Cell Physiol. 2003;285:C334–C342. doi: 10.1152/ajpcell.00043.2003. [DOI] [PubMed] [Google Scholar]
- 18.Luesch H. Mol Biosyst. 2006;2:609–620. doi: 10.1039/b609384a. [DOI] [PubMed] [Google Scholar]
- 19.Chanda SK, White S, Orth AP, Reisdorph R, Miraglia L, Thomas RS, DeJesus P, Mason DE, Huang Q, Vega R, et al. Proc Natl Acad Sci USA. 2003;100:12153–12158. doi: 10.1073/pnas.1934839100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moehlenkamp JD, Johnson JA. Arch Biochem Biophys. 1999;363:98–106. doi: 10.1006/abbi.1998.1046. [DOI] [PubMed] [Google Scholar]
- 21.Hu Q, Klippel A, Muslin AJ, Fantl WJ, Williams LT. Science. 1995;268:100–102. doi: 10.1126/science.7701328. [DOI] [PubMed] [Google Scholar]
- 22.Nakaso K, Kitayama M, Fukuda H, Kimura K, Yanagawa T, Ishii T, Nakashima K, Yamada K. Neurosci Lett. 2000;282:57–60. doi: 10.1016/s0304-3940(00)00836-3. [DOI] [PubMed] [Google Scholar]
- 23.Geetha T, Wooten MW. FEBS Lett. 2002;512:19–24. doi: 10.1016/s0014-5793(02)02286-x. [DOI] [PubMed] [Google Scholar]
- 24.Sanz L, Diaz-Meco MT, Nakano H, Moscat J. EMBO J. 2000;19:1576–1586. doi: 10.1093/emboj/19.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paine MG, Babu JR, Seibenhener ML, Wooten MW. FEBS Lett. 2005;579:5029–5034. doi: 10.1016/j.febslet.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 26.Nakaso K, Yoshimoto Y, Nakano T, Takeshima T, Fukuhara Y, Yasui K, Araga S, Yanagawa T, Ishii T, Nakashima K. Brain Res. 2004;1012:42–51. doi: 10.1016/j.brainres.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 27.Babu JR, Geetha T, Wooten MW. J Neurochem. 2005;94:192–203. doi: 10.1111/j.1471-4159.2005.03181.x. [DOI] [PubMed] [Google Scholar]
- 28.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øverath A, Stenmark H, Johansen T. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimizu S, Narita M, Tsujimoto Y. Nature. 1999;399:483–487. doi: 10.1038/20959. [DOI] [PubMed] [Google Scholar]
- 30.Jordán J, Galindo MF, Tornero D, González-García C, Ceña V. J Neurochem. 2004;89:124–133. doi: 10.1046/j.1471-4159.2003.02299.x. [DOI] [PubMed] [Google Scholar]
- 31.Minagowa N, Kruglov EA, Dranoff JA, Robert ME, Gores GJ, Nathanson MH. J Biol Chem. 2005;280:33637–33644. doi: 10.1074/jbc.M503210200. [DOI] [PubMed] [Google Scholar]
- 32.Khatib ZA, Inaba T, Valentine M, Look AT. Genomics. 1994;23:344–351. doi: 10.1006/geno.1994.1510. [DOI] [PubMed] [Google Scholar]
- 33.Wu L, Liu J, Gao P, Nakamura M, Cao Y, Shen H, Griffin JD. EMBO J. 2005;24:2391–2402. doi: 10.1038/sj.emboj.7600719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komiya T, Park Y, Modi S, Coxon AB, Oh H, Kaye FJ. Oncogene. 2006;25:6128–6132. doi: 10.1038/sj.onc.1209627. [DOI] [PubMed] [Google Scholar]
- 35.Fukasawa K, Fukasawa KM, Kanai M, Fuji S, Hirose J, Harada M. Biochem J. 1998;329:275–282. doi: 10.1042/bj3290275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Šimaga Š, Babíc D, Osmak M, Šprem M, Abramiać M. Gynecol Oncol. 2003;91:194–200. doi: 10.1016/s0090-8258(03)00462-1. [DOI] [PubMed] [Google Scholar]
- 37.Hashimoto J, Yamamoto Y, Kurosawa H, Nishimura K, Hazato T. Biochem Biophys Res Commun. 2000;273:393–397. doi: 10.1006/bbrc.2000.2827. [DOI] [PubMed] [Google Scholar]
- 38.D'Souza I, Schellenberg GD. J Biol Chem. 2006;281:2460–2469. doi: 10.1074/jbc.M505809200. [DOI] [PubMed] [Google Scholar]
- 39.Glatz DC, Rujescu D, Tang Y, Berendt FJ, Hartmann AM, Faltraco F, Rosenberg C, Hulette C, Jellinger K, Hampel H, et al. J Neurochem. 2006;96:635–644. doi: 10.1111/j.1471-4159.2005.03552.x. [DOI] [PubMed] [Google Scholar]
- 40.Marikawa Y, Fujita TC, Alarcón VB. Dev Genes Evol. 2004;214:64–71. doi: 10.1007/s00427-003-0377-x. [DOI] [PubMed] [Google Scholar]
- 41.Lee JM, Moehlenkamp JD, Hanson JM, Johnson JA. Biochem Biophys Res Commun. 2001;280:286–292. doi: 10.1006/bbrc.2000.4106. [DOI] [PubMed] [Google Scholar]
- 42.SantaCruz KS, Yazlovitskaya E, Collins J, Johnson J, DeCarli C. Neurobiol Aging. 2004;25:63–69. doi: 10.1016/s0197-4580(03)00117-9. [DOI] [PubMed] [Google Scholar]
- 43.Li J, Pankratz M, Johnson JA. Toxicol Sci. 2002;69:383–390. doi: 10.1093/toxsci/69.2.383. [DOI] [PubMed] [Google Scholar]
- 44.Borger DR, DeCaprio JA. J Virol. 2006;80:4292–4303. doi: 10.1128/JVI.80.9.4292-4303.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Johnson D, Calkins M, Wright L, Svendsen C, Johnson J. Toxicol Sci. 2005;83:313–328. doi: 10.1093/toxsci/kfi027. [DOI] [PubMed] [Google Scholar]
- 46.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khorova A. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 47.Zitzler J, Link D, Schäfer R, Liebetrau W, Kazinski M, Bonin-Debs A, Behl C, Buckel P, Brinkmann U. Mol Cell Proteomics. 2004;3:834–840. doi: 10.1074/mcp.M400054-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Ishii T, Itoh K, Takahashi S, Sato H, Yanagawa T, Katoh Y, Bannai S, Yamamoto M. J Biol Chem. 2000;275:16023–16029. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- 49.Joung I, Kim HJ, Kwon YK. Biochem Biophys Res Commun. 2005;334:654–660. doi: 10.1016/j.bbrc.2005.06.138. [DOI] [PubMed] [Google Scholar]
- 50.Allard M, Simonnet G, Dupouy B, Vincent JD. J Neurochem. 1987;48:1553–1559. doi: 10.1111/j.1471-4159.1987.tb05700.x. [DOI] [PubMed] [Google Scholar]
- 51.Chen XL, Kunsch C. Curr Pharm Des. 2004;10:879–891. doi: 10.2174/1381612043452901. [DOI] [PubMed] [Google Scholar]
- 52.Lee JM, Li J, Johnson DA, Stein TD, Kraft AD, Calkins MJ, Jakel RJ, Johnson JA. FASEB J. 2005;19:1061–1066. doi: 10.1096/fj.04-2591hyp. [DOI] [PubMed] [Google Scholar]
- 53.Kraft AD, Johnson DA, Johnson JA. J Neurosci. 2004;24:1101–1112. doi: 10.1523/JNEUROSCI.3817-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.