SUMMARY
Identification of novel diagnostic or therapeutic biomarkers from human blood plasma would benefit significantly from quantitative measurements of the proteome constituents over a range of physiological conditions. Herein we describe an initial demonstration of proteome-wide quantitative analysis of human plasma. The approach utilizes post-digestion trypsin-catalyzed 16O/18O peptide labeling, two-dimensional liquid chromatography (LC)-Fourier transform ion cyclotron resonance ((FTICR) mass spectrometry, and the accurate mass and time (AMT) tag strategy to identify and quantify peptides/proteins from complex samples. A peptide accurate mass and LC-elution time AMT tag database was initially generated using tandem mass spectrometry (MS/MS) following extensive multidimensional LC separations to provide the basis for subsequent peptide identifications. The AMT tag database contains >8,000 putative identified peptides, providing 938 confident plasma protein identifications. The quantitative approach was applied without depletion for high abundant proteins for comparative analyses of plasma samples from an individual prior to and 9 h after lipopolysaccharide (LPS) administration. Accurate quantification of changes in protein abundance was demonstrated by both 1:1 labeling of control plasma and the comparison between the plasma samples following LPS administration. A total of 429 distinct plasma proteins were quantified from the comparative analyses and the protein abundances for 25 proteins, including several known inflammatory response mediators, were observed to change significantly following LPS administration.
Keywords: human plasma, proteomics, lipopolysaccharide, 18O labeling, LC-FTICR, accurate mass and time tag
INTRODUCTION
The human plasma proteome has been widely recognized for its significant potential in providing diagnostic or therapeutic biomarkers for various diseases, as well as its potential contribution to personalized medicine.1 As a result, there has been increased interest in comprehensively characterizing the human plasma proteome for the purpose of establishing an extensive database of proteins that could be used for future identification of protein biomarkers indicative of diseases.2-6 Adkins et al. reported the application of a two-dimensional liquid chromatography approach coupled to tandem mass spectrometry (2D-LC-MS/MS) for the analysis of an immunoglobulin-depleted human serum sample, which resulted in the identification of 490 proteins.3 Pieper et al. reported a comprehensive analysis of human serum by using a 3-D whole protein separation process (immunosubtraction/ion exchange/size exclusion) followed by two-dimensional electrophoresis (2DE) and MS identifications of gel spots.2 MS analysis of 1800 gel spots resulted in identification of 325 proteins.2 Recently, we reported on the comprehensive analysis of human plasma and on the qualitative comparison between two different plasma samples using a high resolution 2D-LC-MS/MS approach; both studies resulted in ∼800 plasma protein identifications.4, 7 In addition, the Plasma Proteome Project initiative formed within the Human Proteome Organization (HUPO), is working to obtain a comprehensive analysis of the protein constituents of human plasma and to identify biological sources of variations within individuals over time and across populations.8
While the majority of plasma proteome characterization efforts to date have been qualitative or semi-quantitative, the discovery of novel biomarkers or signature proteins would benefit significantly from quantitative measurements of the differences in plasma protein concentration from different states (e.g., normal vs. diseased states). Recently, several laboratories have reported the applicability of using post-digestion 16O/18O labeling as a quantitative proteomic approach for analysis of complex samples.9-13 In the work reported herein, we describe a global quantitative proteomic approach and its application for comparative analyses of two human plasma samples obtained from a healthy individual prior to (control) and after lipopolysaccharide (LPS) administration (LPS-treated). A 9 h time-point was used in this work only for the initial demonstration of the approach. LPS is a purified bacterial endotoxin known to induce a broad range of inflammatory reactions, including cytokine productions, cell migration, and production of acute-phase proteins14-16. One of our objectives was to identify acute phase plasma proteome changes in response to a prototypical inflammatory-challenge at different time-points (0 h to 24 h) following the LPS administration. Our quantitative proteomic approach combines post-digestion trypsin-catalyzed 16O/18O labeling, strong cation exchange fractionation after the labeling, and LC-Fourier transform ion cyclotron resonance mass spectrometry (FTICR) analyses with the accurate mass and time (AMT) tag strategy11, 17-19 for peptide identification and quantification. This 16O/18O labeling-AMT tag approach was demonstrated to be amenable for high throughput quantitative proteome analyses such as studying the proteomic changes in human plasma following the LPS administration. In a previous initial study, we reported on a qualitative comparison of the two plasma samples following LPS administration based on the number of peptide identifications from LC-MS/MS analyses. Here, we demonstrate more accurate detection of proteomic changes following LPS treatment by using a quantitative approach. Several known inflammatory response or acute-phase mediators were accurately quantified following the administration of LPS.
EXPERIMENTAL PROCEDURES
Human Plasma Sample Preparation
Approval for the conduct of this study was obtained from the Institutional Review Boards of the University of Florida College of Medicine, the Robert Wood Johnson Medical School, the Stanford University School of Medicine, and the Pacific Northwest National Laboratory in accordance with federal regulations.
The human plasma samples were supplied by the Department of Surgery at the University of Florida College of Medicine, which serves as the Sample Collection and Coordination Site for a multicentered clinical study (Inflammation and the Host Response to Injury). The original sample was generated from a healthy adult subject at the Department of Surgery at the Robert Wood Johnson Medical School who, after signed informed consent, received an intravenous injection of Clinical Center Reference Endotoxin (CCRE, Lot 2) LPS (2 ng/kg body weight administered over 5 min). Arterial or venous blood was collected at various time-points between 0 h to 24 h following endotoxin administration. White blood cell counts and various vital signs including body temperature, blood pressure and heat rate were monitored for the subject throughout the 24 h study period. This subject manifested signs and symptoms consistent with those observed after intravenous endotoxin administration to humans20. The plasma samples were prepared from whole blood by centrifugation; samples at T = 0 h (control, baseline immediately prior to endotoxin administration) and T = 9 h (LPS-treated, 9 h following LPS administration) were used for this study. Another set of reference plasma samples obtained from the Stanford University School of Medicine were also used to generate an initial database of peptide identifications.
Aliquots of 200 μL each of the control and LPS-treated plasma samples were diluted and denatured using 8 M urea, 50 mM NH4HCO3, pH 8.2 for 1 h at 37° C and reduced with 10 mM dithiothreitol (DTT) for 30 min at 37° C. Protein cysteinyl residues were alkylated with 40 mM iodoacetamide for 90 min at room temperature, and samples were desalted using a pre-packed PD-10 column containing Sephadex G-25 (Amersham Biosciences, Piscataway, NJ). The protein concentrations for the desalted samples were measured using a BCA protein assay (Pierce, Rockford, IL) that gave total protein amounts of 15.0 mg and 13.9 mg for the control and LPS-treated plasma samples, respectively. The samples were then digested into peptides using sequencing grade trypsin (Promega, Madison, WI) overnight at 37° C with a 1:50 (w:w) trypsin-to-protein ratio. Tryptic activity of residual trypsin was quenched by boiling the samples for 10 min and immediately placing the samples on ice.
Trypsin-catalyzed 16O/18O Labeling
Trypsin-catalyzed 16O/18O labeling was carried out as previously described.11 After residual trypsin activity was quenched via the boiling and quick cooling steps, an aliquot of peptides (1 mg each) was removed from the control and LPS-treated samples, and each aliquot was lyophilized. To dissolve the dried peptides, 40 μL of acetonitrile was first added to the dried digest, followed by the addition of 200 μL of 50 mM NH4HCO3 in either 18O-enriched water (95%, ISOTEC, Miamisburg, OH) or regular 16O water. Then, 2 μL of 1 M CaCl2 and 10 μL of immobilized trypsin (Applied Biosystems, Foster City, CA) were added to the digests and the samples were mixed continuously for 24 h at 30° C. Peptides from the control sample were labeled with 16O and peptides from the LPS-treated samples were labeled with 18O. After labeling, supernatant was collected from each sample after centrifuging the samples for 5 min at 15,000 × g. The corresponding 16O and 18O-labeled samples were pooled, combined, and then lyophilized.
Strong Cation Exchange (SCX) Fractionation
The 16O/18O labeled peptide samples from the control and LPS treated plasma samples were fractionated by SCX similar to that previously described.7, 11 The lyophilized sample was resuspended in 1.5 mL of 10 mM ammonium formate, 25% acetonitrile, pH 3.0, and injected onto a 10 mm × 4.6 mm guard column attached to a Polysulfoethyl A 200 mm × 4.6 mm (5 μm, 300 Å) column (Poly LC, Columbia, MD). The mobile phases consisted of solvent A: 10 mM ammonium formate, 25% acetonitrile, pH 3.0, and solvent B: 500 mM ammonium formate, 25% acetonitrile, pH 6.8. After sample loading, the separation was isocratic for 10 min with 100% solvent A with a flow rate of 1 mL/min. Peptides were eluted using sequential linear gradients from 100% solvent A to 50% solvent B over 40 min and from 50% solvent B to 100% solvent B over another 10 min. The mobile phase was held at 100% solvent B for another 15 min. One mL fractions (1 min per fraction) were collected after the start of the gradient using a Shimadzu FRC-10A fraction collector (Kyoto, Japan) and combined into 30 fractions. Each fraction was lyophilized and analyzed by reversed-phase LC-FTICR.
Reversed-phase Capillary LC-FTICR Analyses
Peptide samples were analyzed using a fully automated custom-built capillary LC system21 coupled online using an in-house-manufactured ESI interface to an Apex III 9.4-Tesla FTICR mass spectrometer (Bruker Daltonics, Billerica, MA). The capillary column was made by slurry packing 5 μm Jupiter C18 bonded particles (Phenomenex, Torrence, CA) into a 65 cm long, 150 μm i.d. fused-silica capillary (Polymicro Technologies, Phoenix, AZ). The mobile phase consisted of 0.2% acetic acid and 0.05% trifluoroacetic acid (TFA) in water (A) and 0.1% TFA in 90% acetonitrile/10% water (B). Mobile phases were degassed on-line using a vacuum degasser (Jones Chromatography Inc., Lakewood, CO). The SCX fractions were dissolved in 50 μL of 25 mM NH4HCO3, pH 8.0. 10 μL aliquots from each peptide sample were injected onto the reversed-phase capillary column for either LC-MS/MS or LC-FTICR analysis. The mobile phase was held at 100% A for 20 min followed by a non-linear exponential gradient elution generated by increasing the mobile-phase composition to ∼70% B over 150 min using a stainless steel mixing chamber. The LC-FTICR mass spectrometer was configured and operated as described elsewhere.22
Generation of a Peptide AMT Tag Database
A database of identified peptides was created based on the results of extensive LC-MS/MS analyses from multiple sample sources. The application of multidimensional LC-MS/MS analyses for profiling two different sets of plasma samples without depletion of any abundant plasma proteins was the same as previously described.4, 7 Human serum albumin and immunoglobulins were removed from the reference plasma sample obtained from Stanford by using a commercial anti-HSA cartridge followed by a Protein G cartridge (Applied Biosystems, Framingham, MA) according to the manufacturer instructions. Both the flow-through following depletion and the eluent were subjected to trypsin digestion, further SCX peptide fractionation, and LC-MS/MS analyses. All LC-MS/MS datasets were analyzed using the SEQUEST algorithm23 (ThermoElectron, San Jose, CA) for peptide and protein identification by searching the MS/MS spectra against the human International Protein Index (IPI) database (consisting of 41,216 protein entries, Version 2.29, April, 2004; available online at http://www.ebi.ac.uk/IPI. A static mass modification on cysteinyl residues that corresponded to alkylation with iodoacetamide (57.0215 Da) was applied during the SEQUEST analysis. The results from all datasets were combined and further filtered for the generation of the AMT tag database and the list of confidently identified peptides.
A set of recently developed filtering criteria24 was applied to filter the data following SEQUEST analyses to generate a list of confidently identified peptides. These criteria are: for the 1+ charge state, Xcorr ≥ 2.0 for fully tryptic peptides and Xcorr ≥ 3.0 for partially tryptic peptides; for the 2+ charge state, Xcorr ≥ 2.4 for fully tryptic peptides and Xcorr ≥ 3.5 for partially tryptic peptides; and for the 3+ charge state, Xcorr ≥ 3.7 for fully tryptic peptides and Xcorr ≥ 4.5 for partially tryptic peptides; ΔCn value of ≥ 0.1 for all charge states. Two additional ΔCn cutoff values 0.05 and 0.15 were applied to reduce false negatives while maintaining the same level of confidence for peptide assignments.24 With the ΔCn value ≥ 0.05, the minimum acceptable Xcorr value was raised to achieve a comparable false positive rate and similarly, for ΔCn valued ≥0.15, the minimum acceptable Xcorr value was reduced. In an attempt to remove redundant protein entries in the reported results, the software program ProteinProphet was used as a clustering tool to group similar or related protein entries into a ‘Protein Group’.25 All peptides that passed the filtering criteria were assigned an identical probability score of 1, and entered into the ProteinProphet program solely for clustering analysis to generate the final non-redundant list of proteins or protein groups.
Peptides that met the same criteria with the exception that ΔCn ≥ 0 were included in the AMT tag database. The peptide retention times from each LC-MS/MS analysis were normalized to a range of 0 to 1 using a predictive peptide LC-normalized elution time (NET) model and linear regression, as previously reported.26 An average NET value and NET standard deviation were assigned to each identified peptide if the same peptide was observed in multiple runs. Both the calculated accurate monoisotopic mass and NET of the identified peptides were included in the AMT tag database.
LC-FTICR Data Analysis
The LC-FTICR datasets were automatically analyzed using in-house software tools that included ICR2LS. The initial analysis of raw LC-FTICR data involved a mass transformation or de-isotoping step using ICR2LS, which is based on the THRASH algorithm.27 The ICR2LS analysis generates a text file report for each LC-FTICR dataset and the report includes both the monoisotopic masses and the corresponding intensities for all detected species for each spectrum. Following ICR2LS analysis, data were processed automatically to yield a two-dimensional mass and LC elution time data set. Data processing steps included filtering data, finding features (i.e., a peak with unique mass and elution time) and pairs of features, computing abundance ratios for pairs of features, normalizing LC elution times, and matching the accurate measured masses and NET values of each feature to the corresponding accurate mass and time (AMT) tag in the database to identify peptide sequences. The peptide sequences of a given feature or pair of features were assigned when the measured mass and NET for each given feature matched the calculated mass and NET of a peptide in the AMT tag database within a 5 ppm mass error and a 5% NET error.
The abundance ratios (18O/16O) for labeled peptide pairs were accurately computed using an equation similar to that previously reported,28
| (1) |
where I0, I2, and I4 are the measured intensities for the monoisotopic peak for a peptide without 18O label, the peak with a mass 2 Da higher than the monoisotopic peak, and the peak with a mass 4 Da higher mass than the monoisotopic peak, respectively. M0, M2, and M4 are the predicted relative abundances for the monoisotopic peak for a peptide, the peak with mass 2 Da higher than the monoisotopic peak, and the peak with mass 4 Da higher than the monoisotopic peak, respectively. The and ratios are estimated using the following two equations (Eq. 2 and 3) according to a recent report,29 where Mr represents the peptide molecular weight.
| (2) |
| (3) |
Ratios from multiple observations of the same peptide across different analyses were averaged to give one ratio per peptide. All quantified peptides were rolled up to non-redundant protein groups using ProteinProphet and the abundance ratio for each protein group was calculated by averaging the ratio of multiple unique peptides stemming from the same protein group.
RESULTS
The Quantitative Proteomic Strategy
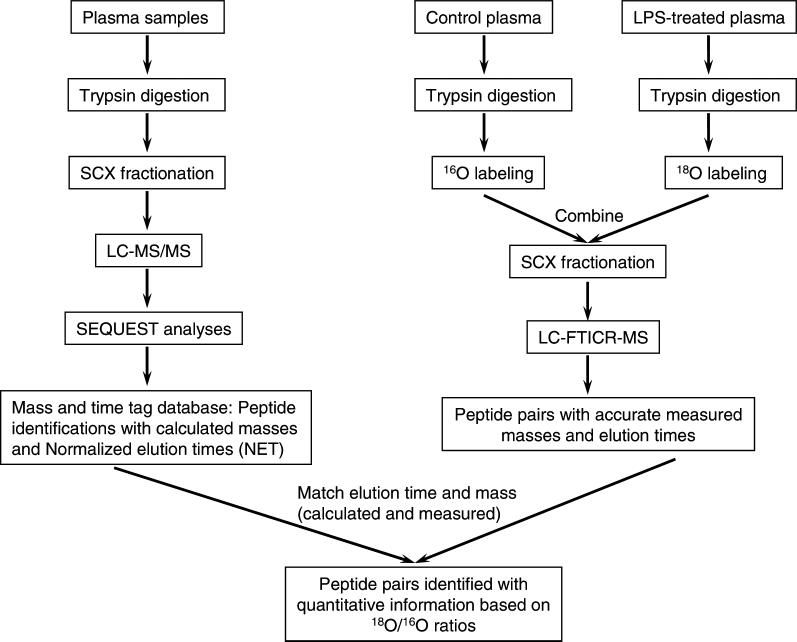
Post-digestion trypsin-catalyzed 18O labeling, SCX fractionation, and the AMT tag approach11, 17-19 were combined to constitute a global quantitative proteomic strategy for analyzing protein abundance changes in human plasma following LPS administration(Figure 1). A similar strategy has been demonstrated using the recently developed quantitative cysteinyl-peptide enrichment technology (QCET).11 The initial stage of generating the peptide AMT tag database was based on extensive LC separations coupled with MS/MS to increase the overall coverage of the proteome. The calculated masses and normalized LC elution times (NET) for all identified peptides were included in the AMT tag database, which then served as a “look-up table” for identifying peptides from LC-FTICR analyses. In the second stage, the control and LPS-treated plasma samples were subjected to trypsin digestion separately. Following digestion, the control sample was labeled with 16O and the treated sample with 18O via the trypsin-catalyzed oxygen exchange reaction. The two labeled samples were then combined for further SCX fractionation to increase the overall dynamic range of the approach. Each SCX fraction was analyzed by LC-FTICR under identical LC conditions, and 16O/18O peptide pairs detected by FTICR were directly identified using the AMT tag database using the mass of the 16O labeled peptide. The 18O/16O ratio of each observed pair provided a precise measurement of the relative abundance for the two versions of the identified peptide, and thus the relative abundances of the peptide in the two samples.
Figure 1.
Strategy for quantitative proteome analysis using 16O/18O labeling and the AMT tag approach. The strategy is a two-stage process. In the initial stage, a peptide AMT tag database for plasma is generated based on extensive analyses of plasma-derived peptide samples using multidimensional LC coupled to tandem mass spectrometry. In the second stage, samples to be compared are separately labeled with 16O and 18O, and then the labeled samples are combined for further SCX fractionation. The fractionated, labeled samples are analyzed by high throughput LC-FTICR and peptide pairs are identified by matching to the AMT tag database and quantified using 18O/16O abundance ratios.
The highly sensitive LC-FTICR measurements provided both (1) sufficient resolution for resolving the 4 Da mass difference 16O/18O labeled pairs with proper deconvolution of the minor overlap for larger peptides, allowing accurate measurements of abundance ratios, and (2) peptide identifications using AMT tags without additional LC-MS/MS analyses, thereby facilitating the high throughput analysis of a large number of samples.
Initial Generation of a Peptide Accurate Mass and Time (AMT) Tag Database for Plasma
To generate a peptide AMT tag database for the human plasma proteome with extensive coverage, we coupled SCX fractionation and capillary LC-MS/MS for the analyses of multiple plasma samples, i.e., the control plasma, LPS-treated plasma, and a reference plasma sample4, 7. Depletion of HSA and immunoglobulin G was performed prior to the 2D-LC-MS/MS analyses to further improve the coverage of low abundance proteins.
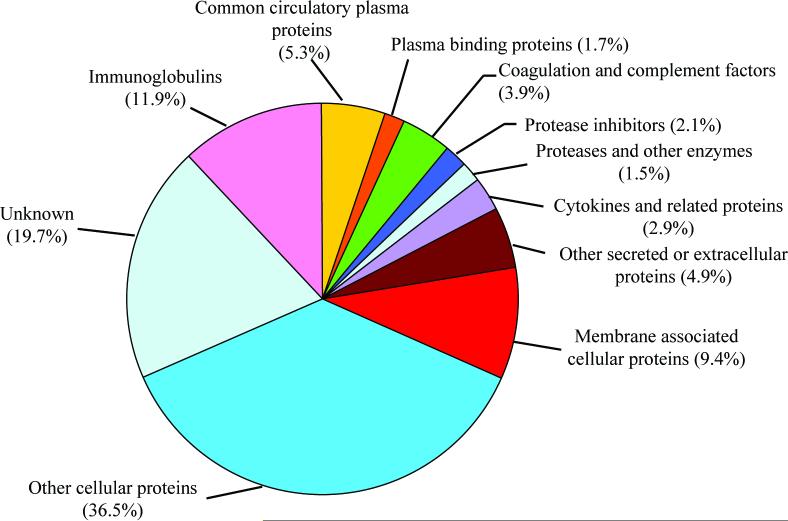
When generating a peptide database to be used for quantitative proteomics, we opted for a conservative approach using only the most confident peptide identifications. The number of peptides and thus proteins identified depends on the stringency of the filtering criteria applied after SEQUEST analyses4. Our recent study using reversed database searching (i.e., analyzing the data against a reversed human protein database with the order of amino acid sequence reversed for each protein, and thus containing a large set of “nonsense” peptides)24 revealed potentially high false positive rates for human plasma/serum peptide identifications based on previously reported criteria3-7. For example, the false positive rate for peptide identifications from human plasma using the criteria described by Washburn et al.30 is ∼30% based upon a reversed database searching24. As a result of our previous study, we developed an improved set of filtering criteria for human plasma that provide a ∼4% false positive rate for peptide identifications 24. Application of these new filtering criteria in our current study resulted in the much more confident identification of a total of 7395 different peptides, covering 938 non-redundant plasma proteins or protein groups. The functional distribution of the 938 proteins is depicted in Figure 2. Approximately 16% of the identified proteins were classified as “classical” plasma proteins (proteins secreted into plasma via mainly the liver and intestines), which includes circulatory and binding proteins, coagulation and complement factors, proteases and inhibitors, cytokines and related proteins. Cellular tissue-derived proteins account for almost half of the total, and a high percentage (20%) of proteins were classified as unknown (either hypothetical, or having no functional annotation). The presence of many tissue-specific cellular proteins in plasma, presumably from cellular “leakage”, further supports the notion that plasma may provide tissue-specific biomarkers; e.g. cancer biomarkers. This complete list of protein/peptide identifications is provided as Supplementary Table 1. To generate the peptide AMT tag database, we applied the same filtering criteria, but with a more liberal ΔCn filter (ΔCn ≥ 0) that allowed peptides with 0 ≤ ΔCn < 0.1 to be also used for peptide identifications when additionally validated by observation at the same LC-NET by FTICR accurate mass measurement. The final AMT tag database contained 8309 unique peptides with accurately measured normalized elution time and calculated monoisotopic mass for each peptide.
Figure 2.
Functional distribution of 938 non-redundant plasma proteins or protein groups (the complete list of identified proteins is available in Supplemental Table 1).
Assessing the Quantitation Accuracy using Labeled Control Plasma Samples
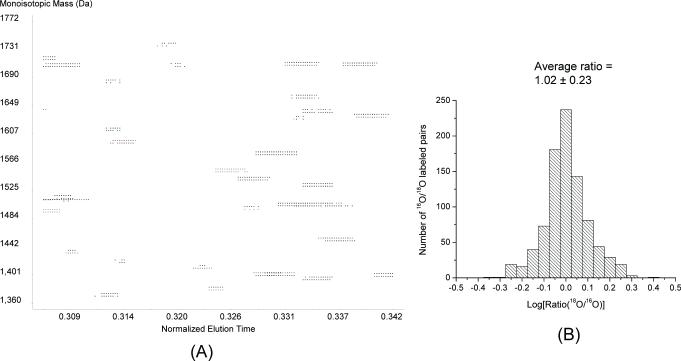
The accuracy of relative quantitation using 16O/18O labeling has been recently demonstrated using standard protein mixtures with known ratios.11 In this work, we further evaluated the labeling and quantitation efficiency by labeling two equal amounts of control plasma samples. For this experiment, two identical aliquots of control plasma were separately digested into peptides and then labeled in either H216O or H218O by trypsin-catalyzed oxygen exchange. After labeling, the samples were combined for LC-FTICR analysis. Figure 3 shows a partial 2D-plot of elution time vs. mass for the detected peptide pairs and the distribution of abundance ratios of all 891 peptide pairs detected from this single LC-FTICR analysis. As shown, the 18O and 16O labeled peptides are readily visualized as co-eluting pairs and the abundance ratio for each peptide's 18O vs. 16O labeled versions can be precisely calculated using Eq. (1). As expected, the distribution of abundance ratios for the detected peptide pairs displayed a Gaussian-like distribution centered at 1.0. The average ratio for all peptide pairs is 1.02 ± 0.23, demonstrating the accuracy for relative abundance measurements even in a complex, proteome-wide sample, and providing the basis in determining changes in relative peptide abundances when using different plasma samples.
Figure 3.
LC-FTICR analysis of 1:1 labeled control plasma samples. (A) A partial 2-D display of the detected 18O/16O labeled peptide pairs. The elution time is shown as a normalized scale between 0 and 1. Observed peaks (represented by spots) correspond to various eluting peptides. The heavy and light isotope-labeled pairs are easily visualized with a 4 Da mass difference. (B) The ratio distribution of 891 detected peptide pairs.
Comparative Analysis of Human Plasma Samples Following LPS Administration.
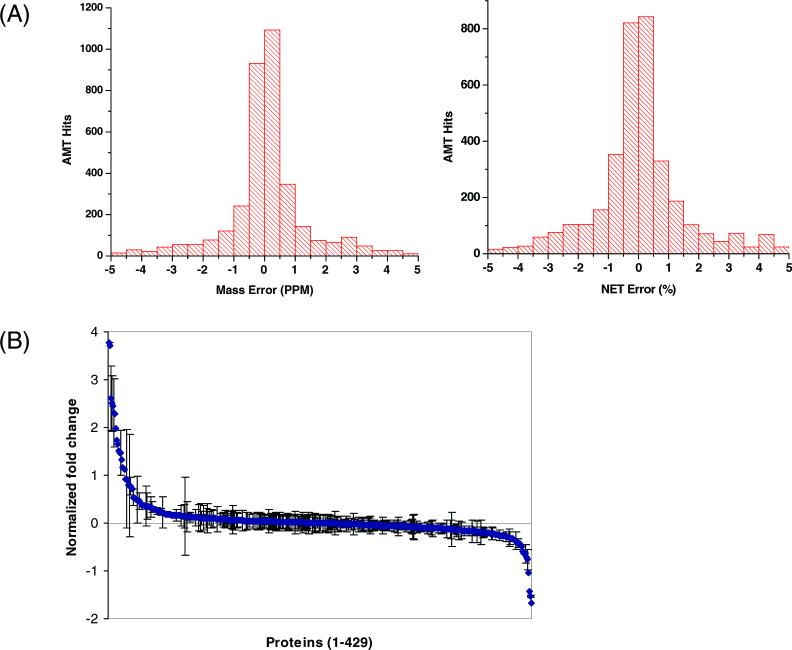
To identify the proteomic changes following LPS administration, plasma samples obtained from a subject before and after LPS administration were labeled with 16O and 18O, respectively. The combined, labeled sample was further separated into 30 fractions by SCX LC, and each fraction was subsequently analyzed by LC-FTICR. A combined total of 3534 different peptide pairs were identified and quantified using a mass tolerance of 5 ppm and NET tolerance of 5%, corresponding to 429 non-redundant plasma proteins. Figures 4A show the distributions of mass error and NET error of the 3534 identified peptide pairs. As shown, the majority of peptide pairs were identified within a mass error of 1 PPM and a NET error of 1%, demonstrating the accuracy of the AMT tag approach for peptide identifications. Figure 4B shows the normalized fold changes of all 429 quantified plasma proteins, demonstrating that the majority of proteins show no significant change, or a minimum change. The standard deviations for protein abundance ratios are shown as error bars in Figure 4B. Relatively small errors (<0.3) were observed for most of the proteins, demonstrating accurate relative quantitation for these proteins. Only a few proteins were observed with large standard deviations in abundance ratios, which indicate poor agreement in abundance ratios of peptides within these proteins. The poor standard deviation observed for these proteins could be due to several reasons: (1) posttranslational modifications may lead to different abundance ratios for different detected peptides from the same protein; (2) the detected peptides may actually correspond to a number of different protein isoforms that were grouped together by ProteinProphet; or (3) an error in assignment due to the small percentage of false peptide identifications.
Figure 4.
(A) The mass error (left) and NET error (right) distributions of the 3534 identified peptide pairs. For those peptide pairs that were observed across multiple fractions, average mass error and NET error values were used in these plots. (B) Normalized fold changes for the 429 quantified proteins following LPS administration. Abundance ratio for each protein shown was normalized to zero (R – 1).35 For ratios smaller than 1, normalized inverted ratios were calculated as [1 – (1/R)]. Error bar for each protein indicates the standard deviation for the abundance ratios from multiple peptides. Proteins without error bars were identified with single peptides.
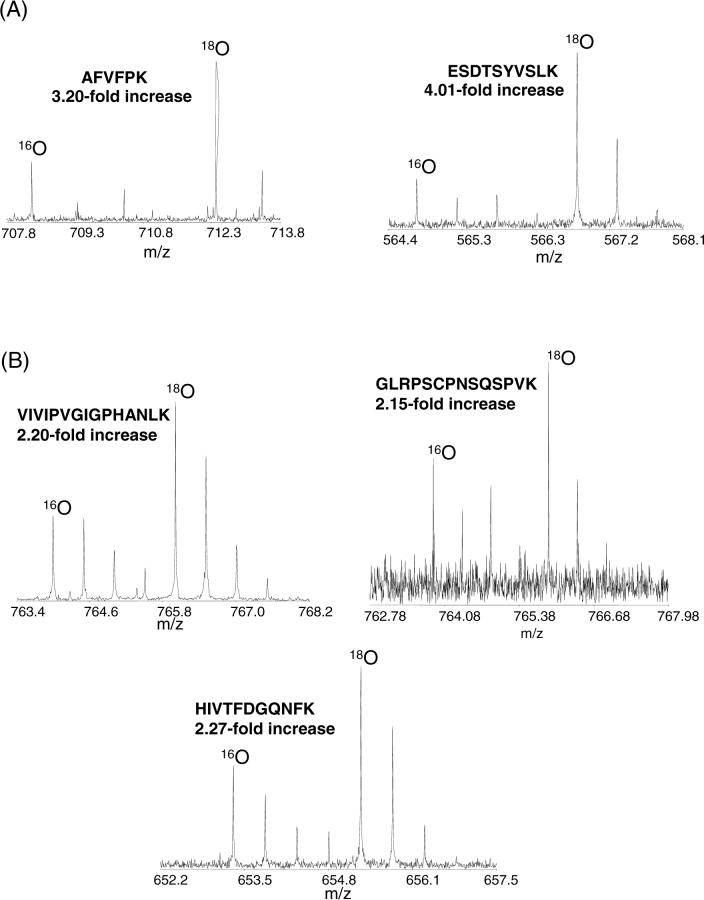
The set of proteins that show significant changes in protein concentration and selected list of proteins that show no significant changes in protein concentration following LPS administration is provided in Table 1. As shown, proteins identified from only a few peptides to those identified with >100 peptides were all quantified with good accuracy, allowing 25 proteins exhibiting significant changes in concentration to be effectively identified. Overall, the quantitative results correlate well with our previous qualitative study.7 Several of the proteins observed with a significant increase in abundance have been previously reported to be inflammatory response markers or mediators whose concentrations are increased during acute phase response. These proteins include C-reactive protein, serum amyloid A, serum amyloid A2, von Willebrand factor, and LPS-binding protein14, 31, 32. The levels of increases in protein abundance for these acute phase proteins are expected for a low level prototypical inflammatory-challenge. Figure 5 shows MS spectra for peptide pairs originating from C-reactive protein and Von Willebrand factor. The different peptide pairs from the same protein show very good agreement in abundance increase. A complete list of the 429 proteins quantified with peptide and protein abundance ratios is available in Supplemental Table 2.
Table 1.
List of selected proteins that were confidently quantified.
| Reference ID | Protein name | # of different peptides quantified | Average 18O/16O abundance ratio | Standard deviation |
|---|---|---|---|---|
| Proteins with significant changes in abundance following LPS administration | ||||
| NP_110381 | Serum amyloid A2 | 1 | 4.78 | |
| SAA_HUMAN | Serum amyloid A | 2 | 4.70 | 0.69 |
| CRP_HUMAN | C-reactive protein | 2 | 3.60 | 0.57 |
| NP_079050 | Hypothetical protein FLJ21294 | 1 | 3.52 | |
| XP_294574 | Similar to A-kinase anchor protein 8 | 2 | 3.45 | 0.71 |
| UTRO_HUMAN | Utrophin | 1 | 3.31 | |
| FAT2_HUMAN | Protocadherin Fat 2 | 1 | 3.29 | |
| CENF_HUMAN | CENP-F kinetochore protein | 1 | 2.98 | |
| NP_060887 | Soluble adenylyl cyclase | 1 | 2.74 | |
| XP_374705 | Similar to polycystic kidney disease 1-like 3 | 1 | 2.65 | |
| CAD5_HUMAN | Vascular endothelial-Cadherin | 1 | 2.50 | |
| VWF_HUMAN | Von Willebrand factor | 5 | 2.47 | 0.47 |
| PSD6_HUMAN | 26s proteasome non-ATPase regulatory subunit 6 | 1 | 2.47 | |
| PRTC_HUMAN | Vitamin K-dependent protein C | 1 | 2.33 | |
| XP_376649 | Hypothetical protein KIAA2005 | 1 | 2.16 | |
| MGA_HUMAN | Maltase-Glucoamylase | 1 | 2.15 | |
| NP_056467 | Hypothetical protein | 1 | 2.12 | |
| HGFL_HUMAN | Hepatocyte growth factor-like protein | 1 | 1.93 | |
| XP_040265 | Hypothetical protein KIAA0217 | 1 | 1.90 | |
| LBP_HUMAN | LPS-binding protein | 4 | 1.78 | 0.18 |
| A2GL_HUMAN | Leucine-rich alpha-2-glycoprotein | 6 | 1.71 | 0.18 |
| GFAP_HUMAN | Glial fibrillary acidic protein, Astrocyte | 1 | 0.37 | |
| OCTC_HUMAN | Peroxisomal carnitine octanoyltransferase | 1 | 0.39 | |
| HBB_HUMAN | Beta globin | 7 | 0.41 | 0.21 |
| CYA1_HUMAN | Brain adenylate cyclase 1 | 1 | 0.49 | |
|
Selected proteins with NO significant changes in abundance following LPS administration | ||||
| CO3_Human | Complement C3 | 137 | 1.01 | 0.18 |
| A2MG_HUMAN | Alpha-2-macroglobulin | 113 | 0.97 | 0.15 |
| TRFE_HUMAN | Serotransferrin | 74 | 0.97 | 0.16 |
| FIBB_HUMAN | Fibrinogen beta chain | 43 | 1.04 | 0.14 |
| CFAB_HUMAN | Complement factor B | 25 | 1.04 | 0.12 |
| PEDF_HUMAN | Pigment epithelium-derived factor | 10 | 1.05 | 0.17 |
| HEP2_HUMAN | Heparin cofactor II | 13 | 1.00 | 0.18 |
| ECM1_HUMAN | Extracellular matrix protein 1 | 7 | 1.02 | 0.15 |
| A2AP_HUMAN | Alpha-2-antiplasmin | 6 | 0.94 | 0.13 |
| TETN_HUMAN | Tetranectin | 5 | 0.96 | 0.14 |
| Q92954 | Megakaryocyte stimulating factor | 3 | 1.28 | 0.06 |
| ALS_HUMAN | Acid labile chain | 3 | 0.93 | 0.09 |
Abundance ratio > 1 indicates an increase in protein concentration following LPS administration. If the abundance ratios differs from 1.0 with more than 3-fold of standard deviation (0.23) from control experiment (either >1.69 or < 0.59), the proteins are considered to be with significant changes in abundance following LPS administration. Proteins with only one unique peptide detected were considered to be less confidently quantified than those proteins with multiple peptides detected.
Figure 5.
Mass spectra for selected peptide pairs. (A) Two different peptide pairs from C-reactive protein. (B) Three different peptide pairs from Von Willebrand factor.
DISCUSSION
Post-digestion trypsin-catalyzed 16O/18O labeling coupled to the AMT tag approach has been applied for global quantitative proteome analysis of human plasma following LPS administration. The results demonstrate that the proteome-wide precise quantitation of different physiological conditions can be achieved using this combined strategy, allowing confident identification of a set of proteins having significant changes in concentration. The coupling of the AMT tag approach with automated data analysis provided the basis for high throughput proteomic measurements without the need for routine LC-MS/MS measurements. The overall efficiency, quantitative precision, and throughput of this strategy make it suitable for comparative proteome analysis of large numbers of clinical samples for biomarker discovery.
There are several advantageous features of the trypsin-catalyzed 16O/18O labeling, including: (1) The post-digestion labeling methodology incorporates 2 atoms of 18O in essentially all tryptic peptides, providing the framework for accurate quantitation; (2) the enzyme catalyzed approach can be applied to label tryptic peptide samples from various biological sources, including tissues, cell lysates, and biological fluids; and (3) the 16O/18O labeling can be easily coupled to peptide-specific enrichment methods such as cysteinyl-peptide enrichment,11 or to peptide-fractionation techniques such as SCX to improve overall proteome coverage of the analysis. However, one of the concerns related 18O labeling is that the 18O-labeled peptides may exchange back to 16O, albeit at a low rate of exchange, that will potentially decrease the overall efficiency of the stable isotope labeling and quantitation. Instead of using cysteine alkylation of trypsin under denaturing conditions as suggested by a previous report13, we found that the residual trypsin activity following digestion can be effectively quenched by boiling the samples for 10 min and then immediately placing the samples on ice. In the post-digestion labeling methodology, the use of immobilized trypsin allows the enzyme to be completely removed following the labeling step. Using this protocol we have not observed any evidence of oxygen back-exchange even after the labeled samples were stored for several months in normal water (data not shown).
We have presented an initial demonstration of the comparative analysis of human plasma without depletion of any major proteins, quantifying a total of 429 non-redundant plasma proteins from clinical human plasma samples. The dynamic range of concentration for proteins present in human plasma is expected to be >1010, which presents a significant challenge for detecting low abundance proteins. While the results from this study demonstrate the effectiveness of identifying proteomic changes between different clinical plasma samples, the limitation in overall detectable dynamic range has resulted in the quantification of proteins that are primarily considered as having a medium to high abundance levels. Thus, the number of proteins observed with significant changes in concentration in plasma is limited by the coverage or low signal levels of lower abundance proteins due to the presence of a limited set of very high abundance proteins. To improve the overall dynamic range of detection, new depletion strategies for removing high abundance proteins2 and novel enrichment methods for enriching specific subsets of peptides such as cysteine-containing peptides11, 33 and N-glycosylated peptides34 are essential and can be applied in combination with the present approach. The application of such depletion or enrichment methodologies is highly promising for extending the present quantitative analysis approach to much larger numbers of low abundance proteins in plasma.
The extension of the reported method to a study of the plasma time-dependent acute phase response following LPS administration is presently in progress that aims to provide an extended dynamic range proteome survey of potential mediators in inflammatory response that may contribute significantly to our understanding of systemic inflammation and sepsis syndrome. With the increased dynamic range of detection resulting from the additional application of depletion and/or enrichment strategies to proteomic samples, we anticipate broad utilization of this quantitative approach in clinical plasma/serum proteomics.
Acknowledgments
We would like to thank Dr. Joshua N. Adkins for critical reviewing the manuscript. The CCRE was kindly supplied by Anthony Suffredini, M.D., NIH Clinical Center, Bethesda, MD. Portions of this research were supported by the National Institute of General Medical Sciences (NIGMS, Large Scale Collaborative Research Grants U54 GM-62119-02), the NIH National Center for Research Resources (RR18522), and the Environmental Molecular Science Laboratory [a national scientific user facility sponsored by the U.S. Department of Energy (DOE) Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory (PNNL)]. PNNL is operated by Battelle Memorial Institute for the DOE under contract DE-AC06-76RLO-1830.
Abbreviations
- LPS
lipopolysaccharide
- SCX
strong cation exchange chromatography
- NET
normalized elution time
- AMT
accurate mass and time
REFERENCES
- 1.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 2.Pieper R, Gatlin CL, Makusky AJ, Russo PS, Schatz CR, Miller SS, Su Q, McGrath AM, Estock MA, Parmar PP, Zhao M, Huang ST, Zhou J, Wang F, Esquer-Blasco R, Anderson NL, Taylor J, Steiner S. The human serum proteome: display of nearly 3700 chromatographically separated protein spots on two-dimensional electrophoresis gels and identification of 325 distinct proteins. Proteomics. 2003;3:1345–1364. doi: 10.1002/pmic.200300449. [DOI] [PubMed] [Google Scholar]
- 3.Adkins JN, Varnum SM, Auberry KJ, Moore RJ, Angell NH, Smith RD, Springer DL, Pounds JG. Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol. Cell Proteomics. 2002;1:947–955. doi: 10.1074/mcp.m200066-mcp200. [DOI] [PubMed] [Google Scholar]
- 4.Shen Y, Jacobs JM, Camp DG, Fang R, Moore RJ, Smith RD, Xiao W, Davis RW, Tompkins RG. High Efficiency SCXLC/RPLC/MS/MS for High Dynamic Range Characterization of the Human Plasma Proteome. Anal. Chem. 2004;76:1134–1144. doi: 10.1021/ac034869m. [DOI] [PubMed] [Google Scholar]
- 5.Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, Conrads TP, Veenstra TD, Adkins JN, Pounds JG, Fagan R, Lobley A. The Human Plasma Proteome: A Nonredundant List Developed by Combination of Four Separate Sources. Mol. Cell Proteomics. 2004;3:311–316. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Tirumalai RS, Chan KC, Prieto DA, Issaq HJ, Conrads TP, Veenstra TD. Characterization of the low molecular weight human serum proteome. Mol. Cell. Proteomics. 2003;2:1096–1103. doi: 10.1074/mcp.M300031-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Qian WJ, Jacobs JM, Camp DG, II, Monroe ME, Moore RJ, Gritsenko MA, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Smith RD. Comparitive proteome analyses of human plasma following lipopolysaccharide treatment using mass spectrometry. Proteomics. 2005;5:572–584. doi: 10.1002/pmic.200400942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Omenn GS. The Human Proteome Organization Plasma Proteome Project pilot phase: reference specimens, technology platform comparisons, and standardized data submissions and analyses. Proteomics. 2004;4:1235–1240. doi: 10.1002/pmic.200300686. [DOI] [PubMed] [Google Scholar]
- 9.Yao X, Afonso C, Fenselau C. Dissection of proteolytic 18O labeling: endoprotease-catalyzed 16O-to-18O exchange of truncated peptide substrates. J. Proteome Res. 2003;2:147–152. doi: 10.1021/pr025572s. [DOI] [PubMed] [Google Scholar]
- 10.Heller M, Mattou H, Menzel C, Yao X. Trypsin catalyzed 16O-to-18O exchange for comparative proteomics: tandem mass spectrometry comparison using MALDI-TOF, ESI-QTOF, and ESI-ion trap mass spectrometers. J. Am. Soc. Mass Spectrom. 2003;14:704–718. doi: 10.1016/S1044-0305(03)00207-1. [DOI] [PubMed] [Google Scholar]
- 11.Liu T, Qian WJ, Strittmatter EF, Camp DG, Anderson GA, Thrall BD, Smith RD. High throughput comparative proteome analysis using a quantitative cysteinyl-peptide enrichment technology. Anal. Chem. 2004;76:5345–5353. doi: 10.1021/ac049485q. [DOI] [PubMed] [Google Scholar]
- 12.Brown KJ, Fenselau C. Investigation of doxorubicin resistance in MCF-7 breast cancer cells using shot-gun comparative proteomics with proteolytic 18O labeling. J. Proteome Res. 2004;3:455–462. doi: 10.1021/pr0340835. [DOI] [PubMed] [Google Scholar]
- 13.Staes A, Demol H, Van Damme J, Martens L, Vandekerckhove J, Gevaert K. Global differential non-gel proteomics by quantitative and stable labeling of tryptic peptides with oxygen-18. J. Proteome Res. 2004;3:786–791. doi: 10.1021/pr049956p. [DOI] [PubMed] [Google Scholar]
- 14.Van Amersfoort ES, Van Berkel TJ, Kuiper J. Receptors, mediators, and mechanisms involved in bacterial sepsis and septic shock. Clin. Microbiol. Rev. 2003;16:379–414. doi: 10.1128/CMR.16.3.379-414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lakhani SA, Bogue CW. Toll-like receptor signaling in sepsis. Curr. Opin. Pediatr. 2003;15:278–282. doi: 10.1097/00008480-200306000-00009. [DOI] [PubMed] [Google Scholar]
- 16.Paludan SR. Synergistic action of pro-inflammatory agents: cellular and molecular aspects. J. Leukoc. Biol. 2000;67:18–25. doi: 10.1002/jlb.67.1.18. [DOI] [PubMed] [Google Scholar]
- 17.Qian WJ, Camp DG, Smith RD. High Throughput Proteomics Using Fourier Transform Ion Cyclotron Resonance (FTICR) Mass Spectrometry. Expert Review of Proteomics. 2004;1:89–97. doi: 10.1586/14789450.1.1.87. [DOI] [PubMed] [Google Scholar]
- 18.Smith RD, Anderson GA, Lipton MS, Pasa-Tolic L, Shen Y, Conrads TP, Veenstra TD, Udseth HR. An Accurate Mass Tag Strategy for Quantitative and High Throughput Proteome Measurements. Proteomics. 2002;2:513–523. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 19.Lipton MS, Pasa-Tolic L, Anderson GA, Anderson DJ, Auberry DL, Battista JR, Daly MJ, Fredrickson J, Hixson KK, Kostandarithes H, Masselon C, Markillie LM, Moore R, Romine MF, Shen Y, Strittmatter E, Tolic N, Udseth HR, Venkateswaran A, Wong KK, Zhao R, Smith RD. Global Analysis of the Deinococcus radiodurans R1 Proteome by Using Accurate Mass Tags. Proc. Natl. Acad. Sci., USA. 2002;99:11,049–011,054. doi: 10.1073/pnas.172170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Poll T, Lowry SF. Biological responses to endotoxin in humans. In: Tellado JM, Forse RA, Solomkin JS, editors. Modulation of the Inflammatory Response in Severe Sepsis. Vol. 20. Karger; Basel: 1995. pp. 18–32. Prog. Surg. [Google Scholar]
- 21.Shen Y, Zhao R, Belov ME, Conrads TP, Anderson GA, Tang K, Pasa-Tolic L, Veenstra TD, Lipton MS, Smith RD. Packed Capillary Reversed-Phase Liquid Chromotaography with High-Performance Electrospray Ionization Fourier Transform Ion Cyclotron Resonance Mass Spectrometry for Proteomics. Anal. Chem. 2001;73:1766–1775. doi: 10.1021/ac0011336. [DOI] [PubMed] [Google Scholar]
- 22.Belov ME, Anderson GA, Wingerd MA, Udseth HR, Tang K, Prior DC, Swanson KR, Buschbach MA, Strittmatter EF, Moore RJ, Smith RD. An automated high performance capillary liquid chromatography-Fourier transform ion cyclotron resonance mass spectrometer for high-throughput proteomics. J. Am. Soc. Mass. Spectrom. 2004;15:212–232. doi: 10.1016/j.jasms.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 24.Qian WJ, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Camp DG, Smith RD. Probability-Based Evaluation of Peptide and Protein Identifications from Tandem Mass Spectrometry and SEQUEST Analysis: The Human Proteome. J. Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 25.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 26.Petritis K, Kangas LJ, Ferguson PL, Anderson GA, Pasa-Tolic L, Lipton MS, Auberry KJ, Strittmatter E, Shen Y, Zhao R, Smith RD. Use of Artificial Neural Networks for the Prediction of Peptide Liquid Chromatography Elution Times in Proteome Analyses. Anal. Chem. 2003;75:1039–1048. doi: 10.1021/ac0205154. [DOI] [PubMed] [Google Scholar]
- 27.Horn DM, Zubarev RA, McLafferty FW. Automated Reduction and Interpretation of High Resolution Electrospray Mass Spectra of Large Molecules. J. Amer. Soc. Mass Spectrom. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 28.Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Protelytic 18O Labeling for Comparative Proteomics: Model Studies with Two Serotypes of Adenovirus. Anal. Chem. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 29.Johnson KL, Muddiman DC. A method for calculating 16O/18O peptide ion ratios for the relative quantification of proteomes. J. Am. Soc. Mass Spectrom. 2004;15:437–445. doi: 10.1016/j.jasms.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 30.Washburn MP, Wolters D, Yates JR. Large-scale Analysis of the Yeast Proteome by Multidimensional Protein Identification Technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 31.Ridker PM, Hennekens CH, Buring JE, Rifai N. C-reactive protein and other markers of inflammation in the prediction of cardiovascular disease in women. N. Engl. J. Med. 2000;342:836–843. doi: 10.1056/NEJM200003233421202. [DOI] [PubMed] [Google Scholar]
- 32.Jensenius M, Ueland T, Fournier PE, Brosstad F, Stylianou E, Vene S, Myrvang B, Raoult D, Aukrust P. Systemic inflammatory responses in African tick-bite fever. J. Infect. Dis. 2003;187:1332–1336. doi: 10.1086/368415. [DOI] [PubMed] [Google Scholar]
- 33.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative Analysis of Complex Protein Mixtures Using Isotope -Coded Affinity Tags. Nat. Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Li X.-j., Martin DB, Aerbersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 2003;21:660–665. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 35.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]