Abstract
Adult hematopoietic stem cells (HSCs) exist in a relatively quiescent state in the bone marrow (BM) microenvironment to fulfill long-term self-renewal and multilineage differentiation functions, an event that is tightly regulated by extrinsic and intrinsic cues. However, the mechanism coordinating the quiescent state of HSCs and their retention in the BM microenvironment remains poorly understood. In a conditional-knockout mouse model, we show that Cdc42−/− HSCs enter the active cell cycle, resulting in significantly increased number and frequency of the stem/progenitor cells in the BM. Cdc42 deficiency also causes impaired adhesion, homing, lodging, and retention of HSCs, leading to massive egress of HSCs from BM to distal organs and peripheral blood and to an engraftment failure. These effects are intrinsic to the HSCs and are associated with deregulated c-Myc, p21Cip1, β1-integrin, and N-cadherin expressions and defective actin organization. Thus, Cdc42 is a critical coordinator of HSC quiescence maintenance and interaction with the BM niche.
Keywords: bone marrow microenvironment, cell cycle, adhesion
Adult hematopoietic stem cells (HSCs) exist in a relatively quiescent state in the bone marrow (BM) microenvironment to execute long-term self-renewal and multilineage differentiation functions (1–3). The maintenance of HSC quiescence involves both extrinsic and intrinsic mechanisms. A number of genes that encode cell cycle or transcriptional regulators, including p21Cip1, p27Kip1, β-catenin/axin, cyclin D1, and c-Myc (4, 5), have been shown to regulate the intrinsic programs of HSCs in this process. In addition, interactions of HSCs with the BM microenvironment in specific anatomical and functional areas, referred to as niches, in the maintenance of HSC quiescence have also gained increasing recognition (6). One hypothesis is that the intrinsic and extrinsic cues, such as bone morphogenic proteins, Ca2+, Notch ligands, and/or Ang-1/Tie2 (7–10), in the BM microenvironment may coordinately regulate the HSC quiescent state.
Despite of the identification of these molecular factors in HSCs and in BM that may collectively contribute to the maintenance of quiescence (7), the mechanism coordinating HSC cell cycle regulation and niche interaction remains unclear. Cdc42 is a ubiquitously expressed member of the Rho GTPase family involved in the regulation of multiple cell functions, including actin polymerization, cell-to-cell or cell-to-extracellular matrix adhesion, and gene transcription (11). Although its function has been extensively studied in various cell systems by expression of dominant negative or constitutively active mutants (12), the physiological roles of Cdc42 in most primary cell lineages, particularly in HSCs, remain unclear. Previously, in a gain-of-Cdc42 activity, Cdc42GAP−/− mouse model, we have found that constitutively increased Cdc42-GTP species cause increased hematopoietic progenitor apoptosis, disorganized actin structure, and defective engraftment without affecting the cell cycle status (13). To further define the role of Cdc42 in HSC regulation, in this study, we present a conditional-knockout mouse model in which the cdc42 gene is inducibly deleted in hematopoietic cells. Our results unveil a role of Cdc42 in maintaining HSC quiescence and in retaining HSCs in the correct location in the BM niche by regulating the expression of a number of key cell cycle regulators (including c-Myc and p21Cip1) and cell adhesion molecules (such as β1-integrin and N-cadherin) and the actin structure.
Results and Discussion
Loss of Cdc42 from HSCs Results in Altered Frequency and Distribution of HSCs.
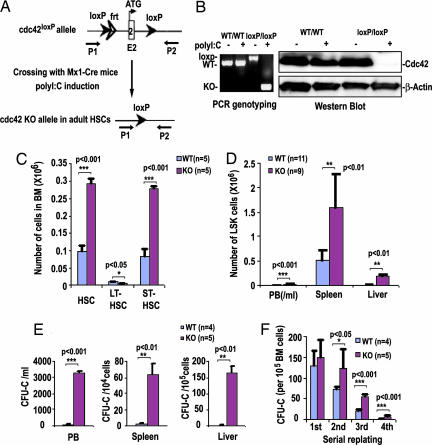
Conventional cdc42 gene-targeted mice die at embryonic day 7.5 (14), precluding a detailed analysis of Cdc42 function in HSCs with this animal model. To circumvent this experimental limitation, we have generated conditional gene-targeted mice with exon 2 of cdc42 gene containing translation initiation codon and nucleotide binding sequences flanked by a pair of loxP sequences [supporting information (SI) Fig. 7]. For examination of the role of Cdc42 in HSC regulation, we cross-bred cdc42loxP/loxP mice with transgenic Mx1-Cre mice to allow IFN-inducible cdc42 gene excision in hematopoietic cells (Fig. 1A). At 5 days after the administration of three doses of poly(inosinic acid)·poly(cytidylic acid) [poly(I·C)] to induce an IFN response in Mx1-Cre;cdc42loxP/loxP mice, the total cellularity of BM did not change (data not shown), but the floxed cdc42 gene sequences and Cdc42 protein became undetectable in the BM cells (Fig. 1B; hereafter referred to as KO).
Fig. 1.
cdc42 gene targeting in hematopoietic cells causes expansion and mobilization of HSCs. (A) Inducible Cre-mediated disruption of cdc42 in Mx1-Cre;cdc42loxP/loxP mice. Arrows indicate the PCR primers P1 and P2. (B) Cdc42 deletion in BM was examined by PCR or anti-Cdc42 Western blotting. (C and D) Numbers of HSCs in BM, including long-term repopulating HSCs (LT-HSCs) and short-term repopulating HSCs (ST-HSCs) (C) and LSK cells in peripheral blood (PB), spleen, and liver (D), were determined by FACS analysis. (E) The colony-forming units in culture activities in PB, spleen, and liver were determined in methylcellulose culture. (F) BM nucleated cells were serially replated in methylcellulose. The numbers of colony-forming units in culture obtained after each round of replating are shown.
To investigate whether Cdc42 deficiency affects HSC function, first we determined the HSC frequency in the BM phenotypically by flow cytometry. Lin−ScaI+c-Kit+ (LSK) cells expressing high or low levels of CD34 were gated (SI Fig. 8) to distinguish between putative long-term repopulating HSCs (LT-HSCs) (Lin−ScaI+c-Kit+CD34low) and short-term repopulating HSCs (Lin−ScaI+c-Kit+CD34high) (15, 16). Loss of Cdc42 led to a 2- to 3-fold increase in phenotypically defined short-term repopulating HSCs and to an ≈2-fold decrease in LT-HSCs (Fig. 1C).
Adult HSCs are normally located in the BM and are mostly absent from peripheral blood (PB) or liver (7–9, 17). Cdc42 deletion led to a drastic increase in the content of LSK population in PB, liver, and spleen, in addition to BM (Fig. 1D). Consistent with these findings, there was a significant increase in the functionally defined progenitor cell numbers in PB, spleen, and liver on cdc42 deletion (Fig. 1E), suggesting that HSCs were mobilized to PB and other tissues by cdc42 deletion. Poly(I:C)-treated WT mice reconstituted with Mx1-Cre;cdc42loxP/loxP BM cells showed similar alterations of tissue distribution of LSK and progenitor cells compared with the KO mice (data not shown), suggesting that the HSC and progenitor mobilization phenotypes are intrinsic to the hematopoietic cells. Serial replating experiments further demonstrated a significantly higher proliferative potential of the KO cells as defined by replating efficiency in colony-forming progenitors (Fig. 1F). These results indicate that Cdc42 deficiency increases the number and alters the distribution of HSCs, leading to a massive mobilization of HSCs and progenitors into peripheral circulation.
Cdc42 Deficiency in HSCs Results in Defective Engraftment.
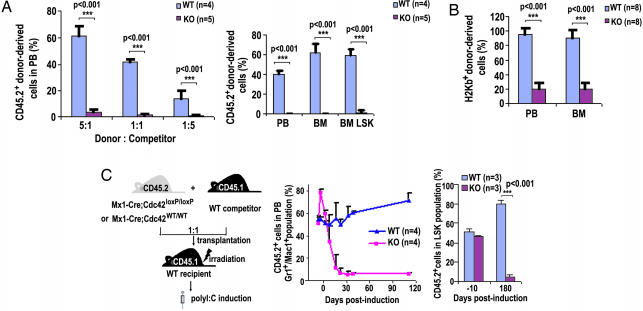
To further assess Cdc42−/− HSC function in vivo, BM cell transplantation into lethally or sublethally irradiated recipient mice was carried out. Competitive repopulation assays combining CD45.1+ WT and CD45.2+ KO BM cells at varying ratios resulted in <5% KO-derived PB or BM cells, whereas the control competitive transplantation using WT cells resulted in the expected genotypic ratios of the PB cells 4 or 10 weeks after transplantation into CD45.1+ BoyJ recipients (Fig. 2A). In a noncompetitive setting, <20% KO donor-derived PB or BM cells, compared with >95% WT donor-derived cells, were detected 8 weeks posttransplantation when sublethally irradiated NOD/SCID mice were used as recipients (Fig. 2B). To rule out potential effects in these assays caused by predeletion of Cdc42, mice engrafted with CD45.2+, Mx1-Cre;cdc42loxP/loxP, or Mx1-Cre;cdc42WT/WT BM cells that were mixed at a 1:1 ratio with CD45.1+ WT competitor BM cells were treated with poly(I·C), and the composition of recipient PB was monitored for up to 180 days (Fig. 2C). The KO Gr1+ and Mac1+ population decreased to <5% after 25 days in these competitive settings (Fig. 2C). The LSK population of the KO genotype in the BM also reduced to <5% 180 days after Cdc42 deletion (Fig. 2C), suggesting that Cdc42 is essential for in vivo HSC engraftment. Thus, Cdc42 deletion leads to an altered stem cell pool that is functionally defective in engraftment and in supporting normal hematopoiesis.
Fig. 2.
Cdc42 deficiency causes an engraftment failure of HSCs. (A) CD45.2+ WT or KO BM nucleated cells were transplanted into lethally irradiated CD45.1+ BoyJ recipient mice at 5:1, 1:1, or 1:5 ratios with CD45.1+ WT competitor cells, and the CD45.2+ cell population in recipients were determined by flow cytometry 20 weeks posttransplantation (Left). The chimerism was also measured in PB, BM, and BM LSK cell populations of 1:1 competitive transplantation recipient mice at the 12 week time point (Right). (B) Sublethally irradiated NOD/SCID recipient mice were used as the transplantation recipients in the absence of competitors. The H2Kb+ donor cells in the PB and BM of recipient mice were determined 8 weeks posttransplantation. (C) The BM cells of the Mx-Cre;cdc42WT/WT or Mx-Cre;cdc42loxP/loxP genotype were transplanted along with WT competitors at a 1:1 ratio into lethally irradiated WT recipients. Eight weeks after reconstitution, mice were treated with poly(I·C) to delete cdc42 as shown in the scheme (Left). The donor-derived Gr1+/Mac1+ lineages in PB (Center) and LSK cells in BM (Right) were quantified at the indicated time points after the poly(I·C) treatment.
Cdc42 Deficiency Leads to HSC Cell Cycle Activation.
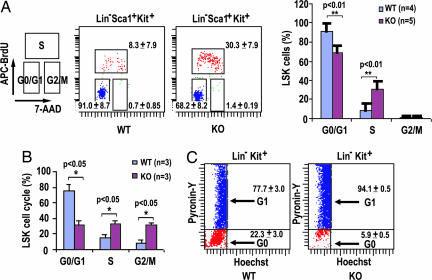
Stem cell loss of function could be related to cell cycle malregulation (2, 4). Because Cdc42 has previously been implicated in cell growth control (11), we tested whether Cdc42−/− HSCs demonstrate a cell cycle or survival defect. To this end, we performed in vivo BrdU labeling in mice to determine the proliferative status of LSK cells in the BM. The percentage of cells in the S phase was significantly higher in Cdc42−/− LSK cells than in WT cells (Fig. 3A). In vitro BrdU pulse-labeling of isolated LSK cells also indicated that Cdc42 deficiency resulted in increased S-phase and G2/M-phase populations (Fig. 3B). To determine whether the enhanced BrdU incorporation in Cdc42−/− LSK cells is associated with an increased stem/progenitor population that is in active cell cycle, we used a combination of DNA and RNA staining by Hoechst 33342 and pyronin Y to distinguish G0 and G1 phases of the cells (2, 4). As shown in Fig. 3C, Cdc42 deficiency caused a significant loss of Lin−c-Kit+ progenitor cells in the G0 phase and an increase in the G1 phase. These cell cycle changes were intrinsic to the HSCs as revealed by an examination of the LSK cells from lethally irradiated transplantation recipients of Mx1-Cre;cdc42loxP/loxP or matching WT donor BM cells after the poly(I·C) treatment (data not shown). In contrast to the cell cycle alterations, however, no difference in apoptosis was detected between KO and WT LSK cells (SI Fig. 9). These results suggest that Cdc42 is required for the maintenance of quiescence, but not survival, of HSCs.
Fig. 3.
Cdc42 deficiency causes cell cycle activation of HSCs. (A) LSK cells in mice were labeled with BrdU in vivo, followed by FACS analysis to assess the cell cycle profile. APC, allophycocyanin; 7-AAD, 7-aminoactinomycin D. Data in the two center panels are displayed as a histogram on the right. (B) Freshly isolated LSK cells were pulse labeled with BrdU for 20 min in vitro. FACS analysis was performed to determine the cell cycle status. Data are representative of three independently performed experiments. (C) BM cells were stained for Lin−c-Kit+ markers and for DNA (Hoechst 33342) and RNA (pyronin Y) and were further analyzed for incorporation of the DNA and RNA dyes to assess the relative proportions in the G0 and G1 phases of the cell cycle.
Cdc42 Deficiency Leads to Homing, Lodging, and Retention Defects of HSCs in the BM Endosteum.
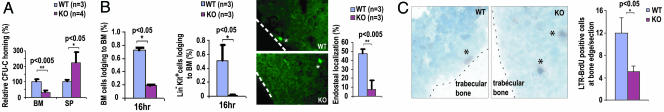
To resolve a potential paradox of the observed cell cycle activation phenotype and the engraftment failure of cdc42−/− HSCs, we next determined whether primitive hematopoietic cells deficient in Cdc42 were able to properly engage the BM niche. Cdc42-deficient progenitors were severely impaired in their ability to enter the BM tissue in a homing assay (Fig. 4A). The lodging ability of fluorescently labeled WT or KO BM or Lin−c-Kit+ BM cells in the BM was further examined by comparing their relative distance from the endosteal surface of BM 16 h after transplantation into nonirradiated recipient mice (10, 18, 19). This analysis revealed a striking reduction of the Cdc42−/− BM and Lin−c-Kit+ BM cells that localized or returned to the endosteal bone surface (Fig. 4B). LT-HSCs, which can retain BrdU labeling over a long period (e.g., 70 days in mice) because of their relative quiescent state, are located close to osteoblastic cells lining the bone surface (8, 9). As shown in Fig. 4C, the number of BrdU long-term retaining (BrdU-LTR) cells at the trabecular bone surface was significantly reduced after cdc42 deletion, suggesting that Cdc42 is also important for LT-HSC retention in the endosteal niche in the BM. These results indicate that Cdc42-deficient HSCs are defective in homing, lodging, and retention in the endosteal niche, which may contribute to the engraftment failure.
Fig. 4.
Cdc42−/− HSCs show defective localization in the BM endosteum. (A) The homing ability of BM cells into an irradiated host was determined. (B) The BM cells (Left) and Lin−c-Kit+ BM cells (Center) lodging into a nonirradiated host were quantified. The spatial distribution of Lin−c-Kit+ BM cells is shown at Right. The stars indicate positive cells in BM, and the dashed lines indicate the margin of bone surface. (C) Immunohistochemical staining of BrdU-LTR cells was performed to reveal its relative localization to the trabecular bone surface (Left). The asterisks indicate BrdU-positive cells in BM, and the dashed lines depict the margin of the bone surface. The relative percentage of BrdU-LTR cells within a 20-cell-diameter distance of the bone surface was quantified (Right).
Cdc42 Is a Critical Regulator of HSC Adhesion, Migration, and Actin Reorganization.
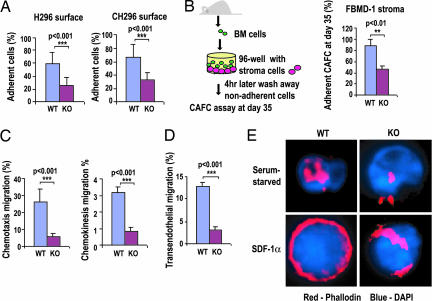
To examine whether the observed homing and lodging defects of KO HSCs are associated with alterations in adhesion activity, adhesion assays of Lin−c-Kit+ progenitor cells to recombinant fibronectin fragments or HSCs to stroma cells were carried out. The adhesion of KO progenitor cells to both fibronectin fragments CH296 (containing both the α4β1 and α5β1 integrin binding sites) and H296 (containing the α4β1 integrin binding site) was significantly reduced (Fig. 5A), as was the adhesion of day 35 cobblestone area-forming cells, which likely represent HSCs, to a BM-derived stroma cell line (FBMD-1) that is capable of supporting hematopoiesis (Fig. 5B). Moreover, the abilities of KO cells to migrate across a transwell, or across an endothelial monolayer, toward a gradient of the chemokine stromal cell-derived factor 1α (SDF-1α), as well as chemokinesis, were severely impaired (Fig. 5 C and D). Isolated KO stem/progenitor cells were also defective in reorganizing F-actin structure on SDF-1α stimulation (Fig. 5E and SI Fig. 10). We conclude that Cdc42 is critical in HSC and progenitor cell adhesion, directional migration, and actin reorganization, functions that are important for HSC homing and retention in the BM microenvironment.
Fig. 5.
Impaired adhesion, migration, and actin organization of stem/progenitor cells on cdc42 deletion. (A) The adhesion activities of BM Lin−c-Kit+ cells to surfaces coated with recombinant fibronectin fragment were compared. (B) The adhesion activities of LT-HSCs (cobblestone area-forming cell day 35) were compared with those of FBMD-1 stroma cells. (C) The chemotaxis migration of BM Lin−c-Kit+ cells in response to an SDF-1α gradient (Left), and the chemokinesis migration of these cells without SDF-1α gradient (Right) are shown. (D) The migration activity of progenitors through a mHEVc murine endothelial cell layer toward a SDF-1α gradient was measured. (E) Isolated Lin−c-Kit+ BM cells were serum-starved and subsequently stimulated with SDF-1α and further stained with rhodamine/phalloidin for actin and DAPI for the nucleus. Images shown are representative of >100 cells examined for each genotype.
Cdc42 Regulates Gene Transcription of Several Key Cell Cycle and Adhesion Regulators in HSCs.
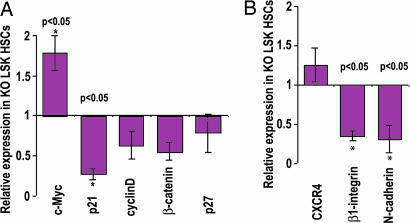
To determine the potential mechanism underlying the accelerated cell cycle status and the defective adhesion properties of the Cdc42 KO HSCs, we further examined the expression profile of a number of cell cycle regulators and adhesion molecules in LSK cells, which have been previously implicated as potential effectors of Rho GTPases and are likely important for HSC quiescent state maintenance or BM retention. Real-time quantitative RT-PCR analysis showed that Cdc42−/− LSK cells expressed a significantly decreased level of p21Cip1 and an increased level of c-Myc compared with WT LSK cells, whereas the cyclin D1 and p27Kip1 mRNA levels remained unchanged (Fig. 6A). Either a decreased level of p21Cip1 or an increased level of c-Myc expression could influence HSC self-renewal and proliferation in the BM microenvironment and may cause a loss of quiescence (4, 20). Real-time quantitative RT-PCR analysis also revealed a significantly decreased expression of β1-integrin and N-cadherin, but not CXCR4, molecules that have been shown to be critical for homing, mobilization, and microlocalization of HSCs (9) in Cdc42-deficient LSK cells (Fig. 6B). These changes may mechanistically contribute to the increased mobilization and decreased interaction of cdc42−/− HSCs with the BM endosteal niche. Taking these findings collectively, it appears that Cdc42 controls the expression of key cell cycle and adhesion regulators and actin structure to coordinate the quiescence maintenance and the BM niche interaction of HSCs.
Fig. 6.
cdc42 deletion altered gene expression of cell cycle and adhesion molecules in HSCs. (A) The mRNA levels of c-Myc, p21Cip1, cyclin D1, β catenin, and p27Kip1 in LSK cells were measured by real-time quantitative RT-PCR. The transcript levels were normalized by using GAPDH of WT cells as an internal control. (B) Relative mRNA transcript levels of CXCR4, β1-integrin, and N-cadherin in LSK cells were measured by real-time quantitative RT-PCR and normalized to the internal GAPDH mRNA transcript of WT cells.
Conclusion
The balance between proliferation and quiescence of HSCs is likely critical in providing long-term, multilineage hematopoiesis in adult life time (21, 22). Because HSCs need to detach from the niche where they are maintained at a quiescent state and migrate to a proliferative zone to enter into asymmetric division and expansion (23), it is believed that HSC interaction with the niche microenvironment and HSC quiescent state maintenance are closely interconnected. The present study shows that deletion of Cdc42 reduces the number and frequency of quiescent HSCs and increases the stem/progenitor population that is actively cycling. Cdc42 deficiency also causes defective homing, lodging, and retention of HSCs in the proper BM niche, which likely results in the impaired engraftment and long-term hematopoiesis. Together with previous characterization of the hematopoietic properties of a Cdc42 gain-of-activity Cdc42GAP knockout mouse model (13), these results suggest that Cdc42 activity represents a critical regulator and coordinator of external and intrinsic cues that control microanatomical location, interaction with the surrounding microenvironment, and cell cycle induction of HSCs.
The current Cdc42 conditional knockout model reveals unique HSC regulatory functions of Cdc42 that are not predictable in the Cdc42 gain-of-activity Cdc42GAP−/− mice (13, 23, 24). Although Cdc42GAP−/− hematopoietic progenitors show normal cell cycle progression but increased apoptosis due to increased JNK activity, Cdc42−/− HSCs display drastically increased cell cycle progression/entry but unaltered survival property. Nonetheless, both Cdc42 gain- and loss-of-activity seems to alter hematopoietic progenitor actin structure and adhesion activity, suggesting that a tightly regulated Cdc42 activity is required for HSC adhesion and migration related functions such as homing, lodging, and engraftment. One aspect of the present findings is that in the absence of Cdc42, LT-HSCs are found dislocated from the “restrictive” niche and move to a relative “proliferation/differentiation-promoting” BM environment (25). The reduced retention in the microenvironment, resulting from actin structure and adhesion defects coupled with altered expression of cell cycle regulatory proteins, such as p21Cip1 and c-Myc, propels Cdc42−/− LT-HSCs to enter into an active cell cycle, giving rise to increased short-term repopulating HSCs and progenitors. These effects appear to be uniquely regulated by Cdc42, because Rac1 and Rac2, two closely related Rho GTPases, are important in HSC retention in the BM niche and for HSC survival and cell cycle progression but are not involved in the maintenance of HSC quiescent state (18, 26), whereas RhoA, another related Rho GTPase, may be involved in HSC engraftment but not retention in the BM niche (12, 27). Whether Cdc42 plays a role in HSC differentiation into various blood cell lineages will be an important question to address in future studies. Another aspect of this study is the implication that Cdc42 function in HSCs is unique, as Cdc42 is known for neuro-stem/progenitor cell polarity establishment and for skin stem/progenitor differentiation into the follicle lineage (28–30) and is required for supporting cell cycle progression through the G1/S phase, rather than maintaining cell cycle quiescence, in mouse embryonic fibroblasts (14, 31). The findings further suggest an avenue for manipulating the HSC cell cycle status as well as the HSC-niche interaction in future therapeutic applications.
Experimental Procedures
Mice.
cdc42loxP/loxP mice were generated in our laboratory by standard recombination procedures, using embryonic stem cells (SI Fig. 7). They were crossbred with Mx1-Cre mice. The protocol for poly(I·C)-induced cdc42 deletion is described in the SI Experimental Procedures.
Transplantation and Engraftment Assays.
The BM transplantation procedures and engraftment assay procedures are described in the SI Experimental Proceedures.
Hematopoietic Progenitor Assays.
The colony-forming units in culture were determined as described in ref. 13. Colonies were scored on day 7, single-cell suspensions were made from pooled colonies, and 1.25 × 105 cells were plated in secondary or tertiary cultures.
Cell Cycle and Survival Analysis.
For assessment of proliferative status of BM cells, mice received single i.p. injections of BrdU (250 mg/kg of body weight). Two hours later, BM cells were harvested and stained for surface markers and then fixed and stained with allophycocyanin-conjugated anti-BrdU antibody by using the Cytofix/Cytoperm Kit (BD Biosciences, San Jose, CA) according to the manufacturer's instructions. Data are representative of three independently performed experiments. BM cells were stained with Hoechst 33342 and pyronin Y at 37°C for 45 min as described in ref. 2. Cells were analyzed by flow cytometry to determine the cell cycle profile. For survival assays, the apoptotic cell population was determined by annexin V staining and analyzed by flow cytometry as described in ref. 13. The numbers are average values ± SD from three mice in each group.
Homing, Lodging, and Spatial Localization Assays of Stem/Progenitor Cells.
Detailed protocols of homing, lodging, and special localization assays of stem progenitor cells are described in the SI Experimental Procedures.
BrdU-LTR Assay.
The BrdU-LTR assay was performed as described in ref. 9. In brief, mice were fed BrdU (0.8 mg/ml) in drinking water for 10 days and injected with poly(I·C) to induce the cdc42 deletion on the 56th, 58th, and 60th day after BrdU labeling. Seventy days after BrdU labeling, immunohistochemical staining of BrdU was performed by using the BrdU In-Situ Detection Kit (BD Biosciences) with the diaminobenzidine substrate according to the manufacturer's instructions.
Gene Expression Analysis.
Real-time quantitative PCR analysis of mRNA from isolated LSK cells is described in the SI Experimental Proceedures.
Supplementary Material
Acknowledgments
We thank Dr. D. A. Williams for a critical reading of the manuscript. This study was supported by National Institutes of Health Grants HL085362 and GM60523 (to Y.Z.).
Abbreviations
- BM
bone marrow
- HSC
hematopoietic stem cells
- LT-HSC
long-term repopulating HSC
- LSK
Lin−ScaI+c-Kit+
- PB
peripheral blood
- poly(I·C)
poly(inosinic acid)·poly(cytidylic acid)
- SDF-1α
stromal cell-derived factor 1α.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610819104/DC1.
References
- 1.Bradford GB, Williams B, Rossi R, Bertoncello I. Exp Hematol. 1997;25:445–453. [PubMed] [Google Scholar]
- 2.Cheshier SH, Morrison SJ, Liao X, Weissman IL. Proc Natl Acad Sci USA. 1999;96:3120–3125. doi: 10.1073/pnas.96.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morrison SJ, Weissman IL. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 4.Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M, Scadden DT. Science. 2000;287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- 5.Lacorazza HD, Yamada T, Liu Y, Miyata Y, Sivina M, Nunes J, Nimer SD. Cancer Cell. 2006;9:175–187. doi: 10.1016/j.ccr.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Xie T. Annu Rev Cell Dev Biol. 2005;21:605–631. doi: 10.1146/annurev.cellbio.21.012704.131525. [DOI] [PubMed] [Google Scholar]
- 7.Arai F, Hirao A, Ohmura M, Sato H, Matsuoka S, Takubo K, Ito K, Koh GY, Suda T. Cell. 2004;118:149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, et al. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Niu C, Ye L, Huang H, He X, Tong WG, Ross J, Haug J, Johnson T, Feng JQ, et al. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 10.Adams GB, Chabner KT, Alley IR, Olson DP, Szczepiorkowski ZM, Poznansky MC, Kos CH, Pollak MR, Brown EM, Scadden DT. Nature. 2005;439:599–603. doi: 10.1038/nature04247. [DOI] [PubMed] [Google Scholar]
- 11.Etienne-Manneville S, Hall A. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Zheng Y. Trends Cell Biol. 2007;17:58–64. doi: 10.1016/j.tcb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Yang L, Filippi MD, Williams DA, Zheng Y. Blood. 2006;107:98–105. doi: 10.1182/blood-2005-05-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen F, Ma L, Parrini MC, Mao X, Lopez M, Wu C, Marks PW, Davidson L, Kwiatkowski DJ, Kirchhausen T, et al. Curr Biol. 2000;10:758–765. doi: 10.1016/s0960-9822(00)00571-6. [DOI] [PubMed] [Google Scholar]
- 15.Goodell MA, Rosenzweig M, Kim H, Marks DF, DeMaria M, Paradis G, Grupp SA, Sieff CA, Mulligan RC, Johnson RP. Nat Med. 1997;3:1337–1345. doi: 10.1038/nm1297-1337. [DOI] [PubMed] [Google Scholar]
- 16.Osawa M, Hanada K, Hamada H, Nakauchi H. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 17.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 18.Cancelas JA, Lee AW, Prabhakar R, Stringer KF, Zheng Y, Williams DA. Nat Med. 2005;11:886–891. doi: 10.1038/nm1274. [DOI] [PubMed] [Google Scholar]
- 19.Wilson A, Trumpp A. Nat Rev Immunol. 2006;6:93–106. doi: 10.1038/nri1779. [DOI] [PubMed] [Google Scholar]
- 20.Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, Pasche AC, Knabenhans C, Macdonald HR, Trumpp A. Genes Dev. 2004;18:2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glimm H, Oh IH, Eaves CJ. Blood. 2000;96:4185–4193. [PubMed] [Google Scholar]
- 22.Passegue E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. J Exp Med. 2005;202:1599–1611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Yang L, Burns K, Kuan CY, Zheng Y. Proc Natl Acad Sci USA. 2005;102:13484–13489. doi: 10.1073/pnas.0504420102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Yang L, Debidda M, Witte D, Zheng Y. Proc Natl Acad Sci USA. 2007;104:1248–1253. doi: 10.1073/pnas.0609149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venezia TA, Merchant AA, Ramos CA, Whitehouse NL, Young AS, Shaw CA, Goodell MA. PLoS Biol. 2004;2:e301. doi: 10.1371/journal.pbio.0020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu Y, Filippi MD, Cancelas JA, Siefring JE, Williams EP, Jasti AC, Harris CE, Lee AW, Prabhakar R, Atkinson SJ, et al. Science. 2003;302:445–449. doi: 10.1126/science.1088485. [DOI] [PubMed] [Google Scholar]
- 27.Ghiaur G, Lee A, Bailey J, Cancelas J, Zheng Y, Williams DA. Blood. 2006;108:2087–2094. doi: 10.1182/blood-2006-02-001560. [DOI] [PubMed] [Google Scholar]
- 28.Chen L, Liao GH, Yang L, Cambell K, Nakafuku M, Kuan CY, Zheng Y. Proc Natl Acad Sci USA. 2006;103:16520–16525. doi: 10.1073/pnas.0603533103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu X, Quondamatteo F, Lefever T, Czuchra A, Meyer H, Chrostek A, Paus R, Langbein L, Brakebusch C. Genes Dev. 2006;20:571–585. doi: 10.1101/gad.361406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cappello S, Attardo A, Wu X, Iwasato T, Itohara S, Wilsch-Brauninger M, Eilken HM, Rieger MA, Schroeder TT, Huttner WB, et al. Nat Neurosci. 2006;9:1099–1107. doi: 10.1038/nn1744. [DOI] [PubMed] [Google Scholar]
- 31.Yang L, Wang L, Zheng Y. Mol Biol Cell. 2006;17:4675–4685. doi: 10.1091/mbc.E06-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.