Abstract
Hepatitis C virus (HCV) and triglyceride-rich very low-density lipoproteins (VLDLs) both are secreted uniquely by hepatocytes and circulate in blood in a complex. Here, we isolated from human hepatoma cells the membrane vesicles in which HCV replicates. These vesicles, which contain the HCV replication complex, are highly enriched in proteins required for VLDL assembly, including apolipoprotein B (apoB), apoE, and microsomal triglyceride transfer protein. In hepatoma cells that constitutively produce infectious HCV, HCV production is reduced by two agents that block VLDL assembly: an inhibitor of microsomal triglyceride transfer protein and siRNA directed against apoB. These results provide a possible explanation for the restriction of HCV production to the liver and suggest new cellular targets for treatment of HCV infection.
Keywords: apolipoprotein B, microsomal triglyceride transfer protein
Many viruses can be produced in large amounts only in certain specialized cell types. A classic example is hepatitis C virus (HCV), a single-stranded positive RNA virus of the Flaviviridae family (1) that can be secreted in abundance only by hepatocytes (2). The factors responsible for this cell type restriction are largely unknown. In the case of HCV, one clue derives from the demonstration that at least a portion of HCV circulates in plasma in complex with very low-density lipoproteins (VLDLs) (3, 4), a family of spherical particles that are produced only in liver (5) to export triglyceride and cholesterol into plasma (6). Although HCV and VLDLs circulate together, a role for VLDLs in viral assembly or secretion has never been demonstrated.
As for all positive-strand RNA viruses, HCV RNA replication occurs in association with cytoplasmic membranes. In the case of HCV, these structures, called “membranous webs,” have been visualized in cultured human hepatoma Huh7 cells that harbor a subgenomic replicon of HCV (7, 8). These replicons are engineered HCV RNA molecules that encode the essential elements for RNA replication, including the nonstructural (NS) proteins NS3, NS4A, NS4B, NS5A, and NS5B (9). After transfection into Huh7 cells, the RNA replicates but does not produce infectious viral particles because it does not encode the structural proteins that are required for assembly and secretion of the virus (9). The membranous webs that harbor the HCV replication complex have never been isolated, and their composition is unknown.
VLDL assembly is currently believed to occur in two different stages, both of which require an enzyme called microsomal triglyceride transfer protein (MTP) (10). In the first stage, MTP transfers triglyceride from cytosolic lipid droplets or the endoplasmic reticulum (ER) to nascent apolipoprotein (apo) B-100, a 540-kDa protein that confers structural integrity to VLDL (11). If sufficient triglyceride is not available, apoB-100 (hereafter referred to as apoB) becomes ubiquitinated and degraded during translation (12). The apoB-containing lipid particles produced in the first stage of VLDL assembly contain only limited amounts of triglyceride (13). In the second stage, apoB-containing VLDL precursor particles fuse with triglyceride droplets in the ER/Golgi luminal compartment (10), a step facilitated by apoE, another major protein component of VLDL (14). Additional triglyceride is transferred by MTP from cytosolic lipid droplets to the luminal compartment in amounts sufficient to promote VLDL assembly in this stage (10). In humans and mice, a genetic defect in MTP severely reduces VLDL secretion (15, 16). Although the first stage of VLDL assembly is known to occur at the ER (13), the exact location of the second stage (i.e., ER, Golgi, or both compartments) is not clear (17).
In these studies, we used a magnetic immuno-isolation procedure (18, 19) to purify membrane vesicles containing the HCV replication complex from Huh7 cells that harbor HCV replicons. We show that these vesicles are highly enriched in apoB, MTP, and apoE, three proteins that participate in the assembly of VLDL. Finally, we demonstrate that agents that inhibit VLDL assembly also inhibit the secretion of HCV from a line of Huh7 cells that constitutively produces infectious HCV. These studies point to a close link between HCV production and VLDL assembly and raise the possibility for treatment of HCV infections with agents that block VLDL secretion.
Results
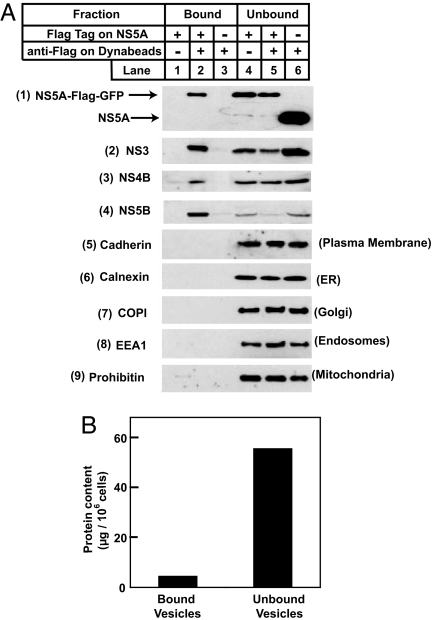
Inasmuch as all HCV NS proteins are localized in a single viral replication complex (7), we undertook to purify the HCV RNA replication complex through use of an antibody directed against an epitope tag attached to the COOH terminus of one of the HCV NS proteins, NS5A, a membrane-associated protein with its COOH terminus facing the cytosol (20). For this purpose, we used Huh7-5A-GFP-6 cells, a line of Huh7 cells harboring a genotype 1b HCV replicon in which a Flag epitope-tagged GFP is inserted at the COOH terminus of NS5A (8). The Flag epitope tag on NS5A renders it possible to use a magnetic immuno-isolation technique to purify membrane vesicles containing NS5A with magnetic Dynabeads coated with anti-Flag. We homogenized the Huh7-5A-GFP-6 cells and incubated the crude homogenates with anti-Flag-coated Dynabeads. As a control, we incubated the homogenates with Dynabeads coated with an irrelevant antibody. As an additional control, we incubated anti-Flag-coated Dynabeads with cell homogenates derived from Huh7-K2040 cells that express a HCV replicon in which NS5A is not epitope-tagged. After magnetic immuno-isolation, bound and unbound vesicles derived from the same amount of cells were lysed with detergent and subjected to SDS/PAGE and immunoblot analysis with antibodies reacting against HCV NS proteins. NS5A was observed in bound vesicles when anti-Flag-coated Dynabeads were incubated with cell homogenates in which NS5A was Flag epitope-tagged (Fig. 1A, blot 1 and lane 2). Approximately 50% of total cellular NS5A was found in the bound vesicles as estimated by densitometry (Fig. 1A, blot 1 and lanes 2 and 5). In the same bound vesicles, we detected ≈50% of NS3, 50% of NS4B, and >80% of NS5B (Fig. 1A, blots 2–4 and lanes 2 and 5). None of the NS proteins was detectable in bound vesicles in the control experiments (Fig. 1A, blots 1–4 and lanes 1 and 3). Bound and unbound vesicles obtained in the above experiment were also subjected to immunoblot analysis with antibodies reacting against markers for plasma membrane, ER, Golgi apparatus, endosomes, and mitochondria. None of these markers was detectable in bound vesicles, even though all of them were easily visible in unbound vesicles (Fig. 1A, blots 5–9). This result indicates that the bound vesicles were not contaminated with major cellular organelles. The total amount of protein in bound vesicles was one-tenth of that in unbound vesicles (Fig. 1B).
Fig. 1.
Localization of HCV NS and cellular proteins in vesicles purified from Huh7 cells harboring HCV replicons. On day 0, Huh7-5A-GFP-6 cells (in which NS5A contains a Flag epitope tag) and Huh7-K2040 cells (in which NS5A is not epitope-tagged) were set up at 7 × 105 cells per 60-mm dish. On day 2, cells were harvested and homogenized. The cell homogenates were incubated with control antibody IgG-2001 or anti-Flag-coated Dynabeads as indicated and subjected to magnetic immuno-isolation as described in Materials and Methods. (A) Lysates of bound and unbound vesicles derived from the same amount of cells (0.05–0.2 per dish) were subjected to SDS/PAGE followed by immunoblot analysis with antibodies reacting against the indicated proteins. Filters were exposed for 10–30 s. (B) Bound and unbound vesicles were solubilized with buffer C, and protein concentration in the detergent lysate was determined by the BCA Protein Assay Kit (Pierce). Each value is the mean of triplicate measurements in the same experiment. Similar results were obtained in more than three independent experiments.
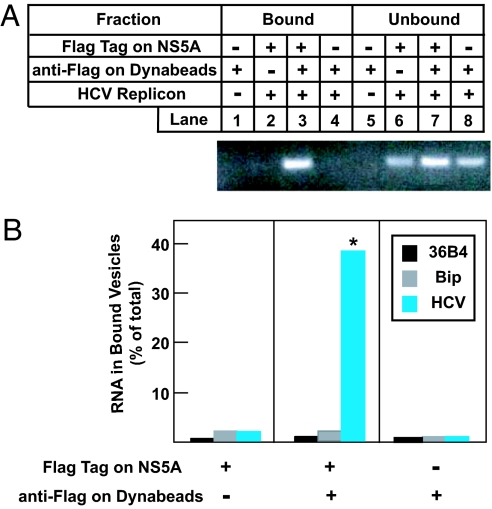
To examine whether the isolated vesicles contained HCV RNA, RNA from bound vesicles and unbound fractions, which include both cytosol and unbound membrane vesicles, was extracted and analyzed by RT-PCR with primers specific for HCV RNA. When unbound fractions were examined, a band of expected size (391 bp) was observed in PCR products derived from Huh7 cells expressing HCV replicons (Fig. 2A, lanes 6–8), but not parental Huh7 cells (Fig. 2A, lane 5). This band was visible only in the PCR product from bound vesicles that were isolated by incubating anti-Flag-coated Dynabeads with cell homogenates containing Flag-tagged NS5A (Fig. 2A, lane 3). We quantified the amount of RNA in bound vesicles and unbound fractions by real-time PCR analysis. As shown in Fig. 2B, 40% of HCV RNA was localized in bound vesicles when anti-Flag-coated Dynabeads were incubated with cell homogenates containing Flag-tagged NS5A. In contrast, these vesicles contained <2% of two cellular RNAs, 36B4 and Bip. No significant amount of viral or cellular RNA was found in bound vesicles in control experiments in which either anti-Flag-coated Dynabeads or the Flag epitope on NS5A were absent. Together with results shown in Fig. 1, these data indicate that the membrane vesicles purified by the magnetic immuno-isolation approach are specifically enriched in HCV NS proteins and RNA, which are characteristic features of the HCV replication complex.
Fig. 2.
Localization of HCV and cellular RNA in vesicles purified from Huh7 cells harboring HCV replicons. On day 0, Huh7, Huh7-5A-GFP-6 cells (in which NS5A contains a Flag epitope tag), and Huh7-K2040 cells (in which NS5A is not epitope-tagged) were set up at 7 × 105 cells per 60-mm dish. On day 2, cells were harvested and homogenized. The cell homogenates were incubated with control antibody IgG-2001 or anti-Flag-coated Dynabeads as indicated and then subjected to magnetic immuno-isolation. RNA was then extracted from bound vesicles derived from two dishes of cells and unbound fractions derived from 1/3 dish of cells and then subjected to RT-PCR analysis with primers specific to HCV RNA (A) and quantitative real-time PCR analysis with the indicated primers (B). Each value in B is the mean of triplicate measurements in the same experiment. An asterisk denotes the level of statistical significance (Student's test) between values obtained from the experiment in which anti-Flag-coated Dynabeads were incubated with cell homogenates containing Flag-tagged NS5A and control experiments in which either anti-Flag-coated Dynabeads or the Flag epitope on NS5A was absent. ∗, P < 0.005. Similar results were obtained in one other independent experiment.
To identify cellular proteins in vesicles containing the HCV replication complex, we performed a large-scale magnetic immuno-isolation by incubating anti-Flag-coated Dynabeads with cell homogenates in which NS5A is Flag-epitope tagged. As a control, we used Dynabeads coated with an irrelevant antibody. Bound vesicles were solubilized by 0.1% Nonidet P-40, a treatment that maintained the attachment of the coated antibody to the Dynabeads. The detergent extracts were then subjected to SDS/PAGE and visualized with colloidal blue [supporting information (SI) Fig. 7]. Segments of the gel that contained visible bands present only in vesicles isolated by anti-Flag but not control antibody-coated Dynabeads were excised and digested with trypsin. The identities of the proteins were determined by tandem MS. This analysis revealed that >30% of the proteins in these vesicles are involved in lipid metabolism (SI Table 1). Two of these proteins, namely apoE and MTP, were chosen for further study as they are both involved in the assembly of VLDL (6). Plasma VLDL is known to associate with clinically isolated HCV particles (3, 4).
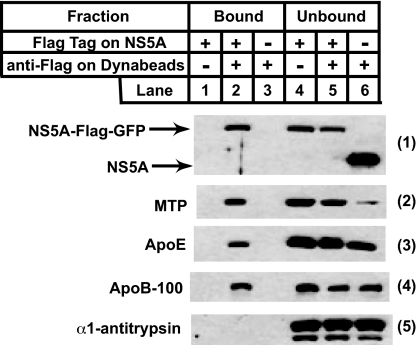
ApoE and apoB are the major protein components of VLDL (5). We did not detect apoB in our MS analysis of bound vesicles, probably because of its high molecular mass (540 kDa) and overall hydrophobicity. To confirm that apoB as well as apoE and MTP are concentrated in vesicles containing the HCV replication complex, these vesicles were isolated as described in Fig. 1 and subjected to immunoblot analysis. Similar to the results shown in Fig. 1, ≈50% of NS5A was observed in bound vesicles when anti-Flag-coated Dynabeads were incubated with cell homogenates in which NS5A was Flag epitope-tagged (Fig. 3, blot 1 and lanes 2 and 5). In the same vesicles we detected ≈50% of MTP, 30% of apoE, and 50% of apoB (Fig. 3, blots 2–4 and lanes 2 and 5). None of these proteins was found in bound vesicles in control experiments in which either anti-Flag-coated Dynabeads or the Flag epitope on NS5A were absent (Fig. 3, blots 2–4 and lanes 1 and 3). α1-Antitrypsin, another abundant protein secreted by hepatocytes (21), was not found in bound vesicles (Fig. 3, blot 5 and lanes 1–3). We detected only the full-length (apoB-100) but not the shorter form of apoB generated by RNA editing (apoB-48) (11) in Huh7-derived cells.
Fig. 3.
Localization of cellular proteins involved in VLDL assembly in vesicles purified from Huh7 cells harboring HCV replicons. Cells were set up, harvested, and analyzed the same way as described in Fig. 1A, except that the immunoblot analysis was performed with antibodies reacting against the indicated proteins. Filters were exposed for 5–30 s.
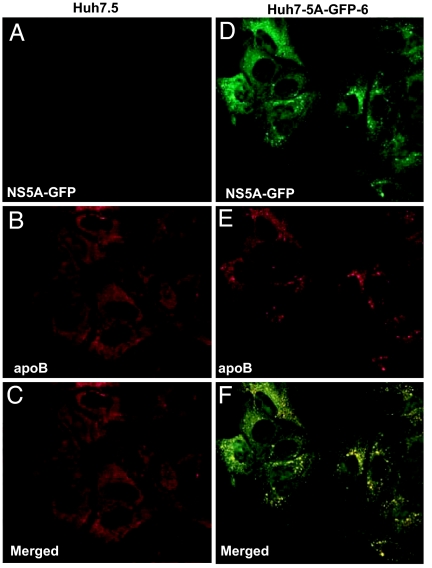
The colocalization of NS5A and apoB in HCV replicon-expressing cells was further investigated by immunofluorescence microscopy. Parental Huh7.5 cells and Huh7-5A-GFP-6 cells were fixed and stained with anti-apoB and anti-GFP to analyze the intracellular distribution of apoB and NS5A, respectively. In the absence of HCV replicons, no NS5A was detected in Huh7.5 cells (Fig. 4A). ApoB in these cells was diffuse throughout the cytoplasm, displaying only a few punctate dots (Fig. 4B). In cells expressing the HCV replicons, much of the NS5A was localized in a punctate pattern consistent with previous studies of the HCV replication complex (22, 23) (Fig. 4D). Most of the apoB in these cells also displayed a punctate staining pattern (Fig. 4E). Merged fluorescent images showed that the vesicles containing apoB colocalized with those containing NS5A (Fig. 4F).
Fig. 4.
Colocalization of apoB with NS5A in Huh7 cells harboring HCV replicons. On day 0, Huh7.5 cells (A–C) and Huh7-5A-GFP-6 (D–F) cells were set up at 1 × 105 cells per well in a six-well plate. On day 2, cells were fixed, stained with anti-GFP (A and D) or anti-apoB (B and E), and subjected to multicolored immunofluorescent microscopy as described in Materials and Methods. (C) A merged fluorescent composite images of A and B. (F) A merged fluorescent composite images of D and E. Within each merged composite image, yellow denotes protein colocalization. (Magnification: ×63.)
To determine whether the vesicles containing the HCV replication complex are derived from the ER or Golgi compartments, we analyzed the susceptibility of N-linked carbohydrate to digestion with endoglycosidase H (endo H), which removes N-linked sugars on proteins localized in ER but not post-Golgi compartments (24). Vesicles containing the HCV replication complex were isolated by the magnetic immuno-isolation approach as described in Fig. 1 and solubilized by detergent. The detergent lysates were treated with endo H or peptide N-glycosidase F (PNGase F), an enzyme that removes N-linked carbohydrates regardless of their subcellular localization (24), and subjected to SDS/PAGE followed by immunoblot analysis with anti-apoB. ApoB in bound vesicles that contain the HCV replication complex was sensitive to both endo H and PNGase F treatments, which caused faster migration of apoB upon electrophoresis (SI Fig. 8, lanes 1–3). The intracellular apoB in unbound vesicles was also endo H-sensitive (SI Fig. 8, lanes 4 and 6). The secreted apoB in the culture medium was resistant to endo H (SI Fig. 8, lanes 7 and 9), but sensitive to the control PNGase F treatment (SI Fig. 8, lane 8). These results suggest that the vesicles containing the HCV replication complex are derived from the ER and that the proteins in the vesicles have not yet reached the Golgi complex.
The observation that apoB, apoE, and MTP were colocalized with HCV RNA replication complexes suggests that HCV RNA replication occurs in a cellular compartment mediating the assembly of VLDL. We thus analyzed whether assembly and secretion of VLDL is required for HCV replication. For this purpose, we used an MTP inhibitor BMS-2101038 that blocks the secretion and accelerates the degradation of apoB by inhibiting MTP-mediated lipid transfer to apoB (25, 26). Huh7-5A-GFP-6 cells were treated with various doses of the MTP inhibitor. To avoid contamination by apoB derived from serum, cells were switched to serum-free medium before harvest. Culture medium was subjected to SDS/PAGE followed by immunoblot analysis. HCV RNA in cell extracts was quantified by real-time PCR analysis. As shown in SI Fig. 9, treatment of cells with as little as 10 nM of the MTP inhibitor completely blocked the secretion of apoB (SI Fig. 9A, upper blot and lane 2). The secretion of α1-antitrypsin was not affected by treatment with the MTP inhibitor (SI Fig. 9A, lower blot). The amount of HCV RNA remained unchanged after cells were treated with up to 100 nM of the MTP inhibitor (SI Fig. 9B). This finding suggests that VLDL secretion is not required for HCV RNA replication.
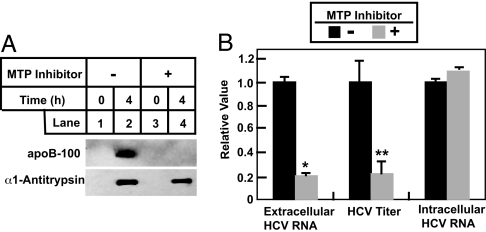
We then examined whether VLDL secretion is required for the release of infectious HCV particles from Huh7 cells. Huh7-GL cells, a line of Huh7 cells that contain a chromosomally integrated genotype 2a HCV cDNA and constitutively produce infectious virus (27), were incubated in the absence or presence of the MTP inhibitor. After incubation in serum-free medium, culture medium was harvested, and the amount of HCV RNA, HCV titer, and apoB in the medium was measured by real-time PCR, foci formation, and immunoblot analysis, respectively. As expected, incubation of cells with the MTP inhibitor blocked the secretion of apoB but not α1-antitrypsin (Fig. 5A). Treatment of the cells with the MTP inhibitor reduced the amount of HCV RNA in the medium and the viral titer by ≈80% (Fig. 5B). The decreased amount of HCV in the medium is not caused by inhibition of HCV RNA synthesis because the amount of intracellular HCV RNA remained the same in the absence or presence of the MTP inhibitor (Fig. 5B). We did not observe an accumulation of intracellular HCV RNA in cells treated with the MTP inhibitor, because even in cells that were not incubated with the inhibitor the amount of HCV RNA detected in the medium was <1% of that found in cells.
Fig. 5.
Decreased secretion of infectious HCV particles from Huh7-GL cells treated with MTP inhibitor. On day 0, Huh7-GL cells were set up at 7 × 105 cells per 60-mm dish. On day 1, cells were treated with (+) or without (−) 100 nM of the MTP inhibitor, BMS-2101038. Sixteen hours later on day 2, cells were switched to serum-free medium in the absence (−) or presence (+) of the same amount of the MTP inhibitor. (A) After incubation for the indicated time, the culture medium was harvested and subjected to SDS/PAGE followed by immunoblot analysis with the indicated antibodies. Filters were exposed for 30 s. (B) After incubation for 4 h, cells and culture medium were harvested. HCV RNA copy numbers and titers in the medium were determined as described in Materials and Methods. Total RNA was prepared from cells and subjected to first-strand cDNA synthesis and quantitative real-time PCR analysis. Values (mean ± SD of three measurements in the same experiment) are plotted relative to the control that was not treated with the MTP inhibitor, which was set at 1. Asterisks denote the level of statistical significance (Student's test) between control cells and cells treated with the MTP inhibitor. ∗, P < 0.005; ∗∗, P < 0.05. HCV copy number and titer in control cells were 7 × 105/ml and 3.5 × 102 foci formation units/ml, respectively. Similar results were obtained in more than five independent experiments.
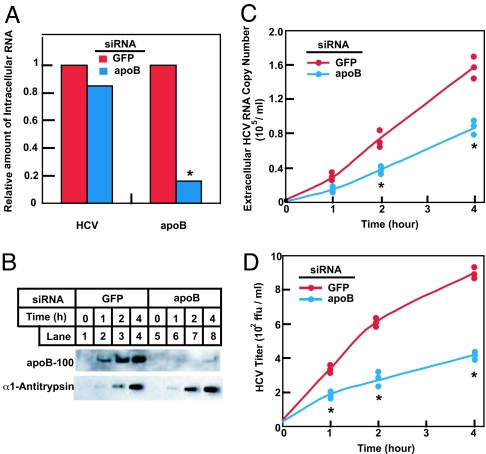
To further confirm that VLDL secretion is required for cellular release of HCV particles, we transfected Huh7-GL cells with a duplex siRNA targeting apoB or GFP as a control. After incubation in serum-free medium, the culture medium was harvested and the amount of apoB and HCV in the medium was analyzed (Fig. 6). Transfection of cells with the apoB siRNA reduced the amount of apoB mRNA by ≈80% without affecting intracellular HCV RNA (Fig. 6A). The apoB siRNA markedly decreased the amount of apoB secreted into the medium, but it did not affect secretion of α1-antitrypsin (Fig. 6B). In control cells transfected with the GFP siRNA, the HCV copy number and titer increased by >10-fold during the 4-h incubation (Fig. 6 C and D). In cells receiving the apoB siRNA, this increase was reduced by ≈50% as assayed by viral copy number (Fig. 6C) and 70% as assayed by the viral titer (Fig. 6D). ApoB RNAi was less potent in inhibiting HCV secretion compared with the MTP inhibitor (Fig. 5) possibly because apoB RNAi did not completely block the secretion of apoB (Fig. 6B, lane 8).
Fig. 6.
Decreased secretion of infectious HCV particles from Huh7-GL cells treated with siRNA targeting apoB. On day 0, Huh7-GL cells were set up at 2 × 105 cells per 60-mm dish. On day 1, cells were transfected with 400 pmol per dish of siRNA duplexes targeting GFP or apoB as indicated. On day 4, cells were switched to serum-free medium. (A) After incubation for 4 h, cells were harvested, after which total RNA was prepared and subjected to first-strand cDNA synthesis and quantitative real-time PCR analysis. Values shown are relative to the GFP siRNA-transfected control, which was set to 1. (B–D) After incubation for the indicated amount of time, culture medium was harvested. (B) The medium was subjected to SDS/PAGE followed by immunoblot analysis with the indicated antibodies. Filters were exposed for 30 s. (C and D) HCV RNA copy number (C) and titer (D) in the medium were determined as described in Materials and Methods. Values from three measurements in the same experiment are shown, each denoted by an individual dot. Asterisks denote the level of statistical significance (Student's test) between GFP siRNA and apoB siRNA-transfected cells. ∗, P < 0.005. Similar results were obtained in one other independent experiment.
Discussion
In these studies we purified membrane vesicles containing the HCV replication complex by a magnetic immuno-isolation approach. The purified vesicles displayed characteristic features of the HCV replication complex in that they were enriched in HCV NS proteins (Fig. 1) and HCV RNA (Fig. 2). The vesicles were highly purified and not contaminated to a significant extent with major cellular organelles (Fig. 1) or abundant cellular mRNAs (Fig. 2). Proteomic analysis of the purified vesicles revealed enrichment for proteins involved in lipid metabolism (SI Table 1). These proteins include apoB, apoE, and MTP, which are proteins required for the assembly of VLDL (5) (Fig. 3). Immunofluorescence microscopy confirmed the colocalization of the HCV replication complex and apoB in vesicular structures (Fig. 4). These vesicular structures were not prominent when we stained control Huh7 cells with anti-apoB, suggesting that these structures may be induced by the viral NS proteins as reported (28). Diversion of nascent VLDL to these structures might explain the previous reports that HCV infection decreases the rate of VLDL secretion in patient (29) and Huh7 cells (30). The vesicles containing the HCV replication complex appears to be derived from the ER because apoB in the vesicles is endo H-sensitive (SI Fig. 8). However, we did not observe an enrichment of calnexin, a general ER marker, in vesicles containing the HCV replication complex (Fig. 1A). Thus, a specialized ER subdomain involved in lipid metabolism and VLDL assembly is most likely to be the organelle where HCV replicates.
The observation that HCV replicates on membrane vesicles assembling VLDL prompted us to investigate the requirement of VLDL synthesis and secretion for the life cycle of HCV. We showed that VLDL synthesis is not required for replication of HCV RNA (SI Fig. 9). This result is consistent with previous findings that HCV RNA can replicate in HeLa and HEK-293 cells (31–33), which do not produce VLDL. The reason for colocalization of the HCV replication and VLDL assembly appears to lie in a requirement for coassembly or secretion of VLDL and HCV particles. Thus, treatment of cells with an MTP inhibitor or siRNA targeting apoB that specifically blocks the secretion of VLDL inhibits HCV production at the same time (Figs. 5 and 6). This finding, together with previous reports showing that clinically isolated HCV particles are associated with VLDL (3, 4), suggests that HCV particles are attached to or incorporated into VLDL during the assembly of the lipoprotein particles and secreted together with VLDL. In experiments not shown, we were unable to demonstrate an association between secreted HCV and VLDL in our tissue culture system by using density gradient ultracentrifugation (apoB was found in fractions with d < 1.01 g/ml in these experiments) or coimmunoprecipitation to isolate the complex. It is possible that the HCV·VLDL complex produced in cultured cells is much more fragile than that isolated from infected patients so that the complex is disrupted during centrifugation.
HCV production was inhibited when cells were treated with the MTP inhibitor (Fig. 5). However, such inhibition was not complete, and ≈20% of HCV was still released into the medium (Fig. 5B). Thus, it is possible that some HCV particles may be secreted out of cells in a VLDL-independent manner. An alternative possibility is that a tiny amount of secreted VLDL beyond the detection limit of immunoblot analysis is enough to mediate the release of the small percentage of HCV.
Besides liver, apoB-containing triglyceride-rich lipoproteins are also produced in intestine (6), which is not known to produce HCV in large quantity. These lipoproteins, called chylomicrons, contain a shorter form of apoB (apoB-48) and their assembly is not exactly the same as that of VLDL produced in liver (34). This difference may prevent HCV from secreting together with chylomicrons. If this is the case, proteins mediating the assembly of VLDL may be the liver-specific cellular factors that support HCV infection. Inasmuch as continued viral production is believed to be necessary for sustained viral infection (2), drugs that block the assembly and secretion of VLDL may represent cellular targets for treatment of HCV infection. In this regard, siRNA targeting hepatic apoB inhibited VLDL secretion in mice (35) and primates (36), but it has not yet been tested in humans. Antisense RNA drugs targeting apoB (37) and several MTP inhibitors have already been tested in clinical trails because of their ability to block VLDL secretion, thereby lowering the level of plasma triglyceride and LDL cholesterol (38, 39). Long-term treatment with MTP inhibitors led to the accumulation of fat in livers (39). However, short-term treatment (up to several weeks) reduced the level of VLDL with only minor adverse effects, which disappeared after drug removal (39). It will be interesting to examine whether short-term treatment with MTP inhibitors is beneficial in treating HCV infection.
Materials and Methods
Descriptions of RNA interference, immunoblot analysis, real-time PCR, HCV titer, and immunofluorescent microscopy are in SI Text.
Materials.
We obtained Dynabeads M-500 Subcellular from Invitrogen (Carlsbad, CA) and endo H and PNGase F from New England Biolabs (Ipswich, MA). Antibody reagents were obtained from the following sources: goat anti-mouse IgG and mouse monoclonal anti-Flag tag M2 antibody from Sigma (St. Louis, MO); rabbit polyclonal anti-calnexin, anti-EEA1, anti-prohibitin, anti-pan cadherin, and anti-GFP from Abcam (Cambridge, MA); goat polyclonal anti-NS3 from USBiological (Swampscott, MA); goat polyclonal anti-apoE from CalBiochem (San Diego, CA); sheep polyclonal anti-apoB and anti-α1-antitrypsin from Biodesign International (Kennebunkport, ME); rabbit polyclonal anti-GFP from Invitrogen; HPR-conjugated donkey anti-mouse, anti-rabbit, anti-sheep, and anti-goat IgGs (affinity-purified) from Jackson Immunoresearch Laboratories (West Grove, PA); rabbit polyclonal anti-NS4B from Ralf Bartenschlager (University of Heidelberg, Heidelberg, Germany); mouse monoclonal anti-NS5B from Guangxiang Luo (University of Kentucky College of Medicine, Lexington, KY); rabbit polyclonal anti-COPI from Joachim Seemann (University of Texas Southwestern Medical Center); and rabbit polyclonal anti-MTP from Larry Swift (Vanderbilt University, Nashville, TN). Human anti-HCV serum (40) and control mouse mAb IgG-2001 (41) have been described. Mouse monoclonal anti-apoB (1D1) has been described (42) and was provided by Jay Horton (University of Texas Southwestern Medical Center). Polyclonal antibody against NS5A was produced by immunizing rabbits with purified full-length NS5A as described (43). The MTP inhibitor BMS-2101038 (25) was synthesized by Chuo Chen (University of Texas Southwestern Medical Center).
Cell Culture.
Huh7 cells and Huh7.5 cells, a mutant line of Huh7 cells that support HCV replication in high efficiency (44), were maintained in medium A (DMEM with 4.5 g/liter glucose, 100 units/ml penicillin, 100 μg/ml streptomycin sulfate, and 10% FCS). Huh7-K2040 cells (22) and Huh7-5A-GFP-6 cells (8) are Huh7 cells that harbor genotype 1b HCV subgenomic replicons. They were maintained in medium A supplemented with 200 μg/ml G418. Huh7-GL cells, a line of Huh7 cells that contain a chromosomally integrated genotype 2a HCV cDNA and constitutively produce infectious virus (27), were maintained in medium A supplemented with 5 μg/ml blasticidine. All cells were maintained in monolayer culture at 37°C in 5% CO2. Huh7.5 and Huh7-5A-GFP-6 cells were obtained from Charles Rice (The Rockefeller University, New York, NY), whereas Huh7-GL cells were a gift from Guangxiang Luo.
Magnetic Immuno-Isolation of Membrane Vesicles.
All operations were performed at 4°C. Dynabeads M-500 were coated with control IgG-2001 or anti-Flag according to the manufacturer's protocol. For small-scale isolation suitable for immunoblot analysis, cells pooled from two dishes (60 mm) were harvested and resuspended in 400 μl of buffer A [50 mM Hepes-KOH, pH 7.2/0.25 M sorbitol/0.25 M potassium acetate/10% (vol/vol) FCS/2 mM sodium EDTA/10 μg/ml leupeptin/5 μg/ml pepstatin A] and homogenized by passing through a 22.5-gauge needle 30 times. Each cell homogenate received a direct addition of 800 μl of buffer A followed by centrifugation at 500 × g for 5 min. The resulting supernatant was mixed with 25 μl of antibody-coated Dynabeads M-500. After rotating at 4°C for 16 h, the reaction mixture was placed on a magnetic stand (Invitrogen). The unbound homogenate (designated as unbound fraction) was centrifuged at 100,000 × g for 30 min, and the pellet was designated as unbound vesicles. Dynabeads with bound vesicles were washed four times (10 min each) with 1 ml of buffer B (50 mM Hepes-KOH, pH 7.2/0.25 M sorbitol/0.25 M potassium acetate) supplemented with 1 mg/ml BSA, followed by an additional wash with 1 ml of buffer B. Vesicles remaining bound to Dynabeads were designated as bound vesicles. For large-scale isolation required for proteomic analysis, cells pooled from 50 dishes (100 mm) were harvested and resuspended in 4 ml of buffer A and homogenized as described above. Each cell homogenate received an addition of 4 ml of buffer A followed by centrifugation at 500 × g for 5 min. The resulting supernatant was mixed with 1 ml of antibody-coated Dynabeads M-500, and vesicles were isolated as described above. Bound vesicles were lysed with 500 μl of buffer C [50 mM Tris·HCl, pH 7.2/150 mM NaCl/0.1% (vol/vol) Nonidet P-40/10 μg/ml leupeptin/5 μg/ml pepstatin A]. Each lysate was then concentrated to 50 μl by using microcon YM-3 (Millipore, Billerica, MA) and subjected to 10% SDS/PAGE followed by colloidal blue staining. Selected bands were excised, and the identities of the proteins were determined by tandem MS performed by the Protein Identification Service at the University of Texas Southwestern Medical Center.
Supplementary Material
Acknowledgments
We thank Michael S. Brown and Joseph L. Goldstein for their constant support and helpful comments, Saada Abdalla for excellent technical assistance, Lisa Beatty and Marissa Hodgin for invaluable assistance in tissue culture, Jeff Cormier for real-time PCR analysis, and the Protein Identification Service at the University of Texas Southwestern Medical Center for assistance in proteomic analysis. This work was supported by National Institutes of Health Grants HL-20948 and AI-060389 and the Perot Family Foundation.
Abbreviations
- HCV
hepatitis C virus
- MTP
microsomal triglyceride transfer protein
- VLDL
very low-density lipoproteins
- apo
apolipoprotein
- ER
endoplasmic reticulum
- NS
nonstructural
- endo H
endoglycosidase H.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700760104/DC1.
References
- 1.Appel N, Schaller T, Penin F, Bartenschlager R. J Biol Chem. 2006;281:9833–9836. doi: 10.1074/jbc.R500026200. [DOI] [PubMed] [Google Scholar]
- 2.Chisari FV. Nature. 2005;436:930–932. doi: 10.1038/nature04076. [DOI] [PubMed] [Google Scholar]
- 3.Andre P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Brechot C, Paranhos-Baccala G, Lotteau V. J Virol. 2002;76:6919–6928. doi: 10.1128/JVI.76.14.6919-6928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen SU, Bassendine MF, Burt AD, Martin C, Pumeechockchai W, Toms GL. J Virol. 2006;80:2418–2428. doi: 10.1128/JVI.80.5.2418-2428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibbons GF, Wiggins D, Brown A-M, Hebbachi A-M. Biochem Soc Trans. 2004;32:59–64. doi: 10.1042/bst0320059. [DOI] [PubMed] [Google Scholar]
- 6.Gibbons GF, Islam K, Pease RJ. Biochim Biophys Acta. 2000;1483:37–57. doi: 10.1016/s1388-1981(99)00182-1. [DOI] [PubMed] [Google Scholar]
- 7.Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. J Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moradpour D, Evans MJ, Gosert R, Yuan Z, Blum HE, Goff SP, Lindenbach BD, Rice CM. J Virol. 2004;78:7400–7409. doi: 10.1128/JVI.78.14.7400-7409.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lohmann V, Körner F, Koch JO, Herian U, Theilmann L, Bartenschlager R. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 10.Shelness GS, Sellers JA. Curr Opin Lipidol. 2001;12:151–157. doi: 10.1097/00041433-200104000-00008. [DOI] [PubMed] [Google Scholar]
- 11.Olofsson SO, Boren J. J Intern Med. 2005;258:395–410. doi: 10.1111/j.1365-2796.2005.01556.x. [DOI] [PubMed] [Google Scholar]
- 12.Avramoglu RK, Adeli K. Rev Endocrine Metab Disord. 2004;5:293–301. doi: 10.1023/B:REMD.0000045100.66675.92. [DOI] [PubMed] [Google Scholar]
- 13.Gusarova V, Brodsky JL, Fisher EA. J Biol Chem. 2003;278:48051–48058. doi: 10.1074/jbc.M306898200. [DOI] [PubMed] [Google Scholar]
- 14.Mensenkamp AR, Havekes LM, Romijn JA, Kuipers F. J Hepatol. 2001;35:816–822. doi: 10.1016/s0168-8278(01)00249-5. [DOI] [PubMed] [Google Scholar]
- 15.Sharp D, Blinderman L, Combs KA, Kienzle B, Ricci B, Wager-Smith K, Gil CM, Turck CW, Boumas ME, Rader DJ, et al. Nature. 1993;365:65–69. doi: 10.1038/365065a0. [DOI] [PubMed] [Google Scholar]
- 16.Raabe M, Flynn LM, Zlot CH, Wong JS, Veniant MM, Hamilton RL, Young SG. Proc Natl Acad Sci USA. 1998;95:8686–8691. doi: 10.1073/pnas.95.15.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher EA, Ginsberg HN. J Biol Chem. 2002;277:17377–17380. doi: 10.1074/jbc.R100068200. [DOI] [PubMed] [Google Scholar]
- 18.Rowe T, Aridor M, McCaffery JM, Plutner H, Nuoffer C, Balch WE. J Cell Biol. 1996;135:895–911. doi: 10.1083/jcb.135.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Randhawa VK, Bilan PJ, Khayat ZA, Daneman N, Liu Z, Ramlal T, Volchuk A, Peng XR, Coppola T, Regazzi R, et al. Mol Biol Cell. 2000;11:2403–2417. doi: 10.1091/mbc.11.7.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brass V, Bieck E, Montserret R, Wolk B, Hellings JA, Blum HE, Penin F, Moradpour D. J Biol Chem. 2002;277:8130–8139. doi: 10.1074/jbc.M111289200. [DOI] [PubMed] [Google Scholar]
- 21.Lomas DA, LI-Evans D, Finch JT, Carrell RW. Nature. 1992;357:605–607. doi: 10.1038/357605a0. [DOI] [PubMed] [Google Scholar]
- 22.Ye J, Wang C, Sumpter R, Jr, Brown MS, Goldstein JL, Gale M., Jr Proc Natl Acad Sci USA. 2003;100:15865–15870. doi: 10.1073/pnas.2237238100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Hage N, Luo G. J Gen Virol. 2003;84:2761–2769. doi: 10.1099/vir.0.19305-0. [DOI] [PubMed] [Google Scholar]
- 24.Kornfeld R, Kornfeld S. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 25.Wetterau JR, Gregg RE, Harrity TW, Arbeeny C, Cap M, Connolly F, Chu CH, George RJ, Gordon DA, Jamil H, et al. Science. 1998;282:751–754. doi: 10.1126/science.282.5389.751. [DOI] [PubMed] [Google Scholar]
- 26.Higashi Y, Itabe H, Fukase H, Mori M, Fujimoto Y, Takano T. J Biol Chem. 2003;278:21450–21458. doi: 10.1074/jbc.M301376200. [DOI] [PubMed] [Google Scholar]
- 27.Cai Z, Zhang C, Chang KS, Jiang J, Ahn BC, Wakita T, Liang TJ, Luo G. J Virol. 2005;79:13963–13973. doi: 10.1128/JVI.79.22.13963-13973.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serfaty L, Andreani T, Giral P, Carbonell N, Chazouilleres O, Poupon R. J Hepatol. 2001;34:428–434. doi: 10.1016/s0168-8278(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 30.Domitrovich AM, Felmlee DJ, Siddiqui A. J Biol Chem. 2005;280:39802–39808. doi: 10.1074/jbc.M510391200. [DOI] [PubMed] [Google Scholar]
- 31.Kato T, Date T, Miyamoto M, Zhao Z, Mizokami M, Wakita T. J Virol. 2005;79:592–596. doi: 10.1128/JVI.79.1.592-596.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ali S, Pellerin C, Lamarre D, Kukolj G. J Virol. 2004;78:491–501. doi: 10.1128/JVI.78.1.491-501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu Q, Guo JT, Seeger C. J Virol. 2003;77:9204–9210. doi: 10.1128/JVI.77.17.9204-9210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahmood Hussain M. Atherosclerosis. 2000;148:1–15. doi: 10.1016/s0021-9150(99)00397-4. [DOI] [PubMed] [Google Scholar]
- 35.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, et al. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 36.Zimmermann TS, Lee ACH, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, et al. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 37.Burnett JR. Curr Opin Mol Ther. 2006;8:461–467. [PubMed] [Google Scholar]
- 38.Cuchel M, Bloedon LT, Szapary PO, Kolansky DM, Wolfe ML, Sarkis A, Millar JS, Ikewaki K, Siegelman ES, Gregg RE, et al. N Engl J Med. 2007;356:148–156. doi: 10.1056/NEJMoa061189. [DOI] [PubMed] [Google Scholar]
- 39.Chandler CE, Wilder DE, Pettini JL, Savoy YE, Petras SF, Chang G, Vincent J, Harwood HJ., Jr J Lipid Res. 2003;44:1887–1901. doi: 10.1194/jlr.M300094-JLR200. [DOI] [PubMed] [Google Scholar]
- 40.Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, et al. Proc Natl Acad Sci USA. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tolleshaug H, Goldstein JL, Schneider WJ, Brown MS. Cell. 1982;30:715–724. doi: 10.1016/0092-8674(82)90276-8. [DOI] [PubMed] [Google Scholar]
- 42.Milne RW, Theolis R, Verdery RB, Marcel YL. Arteriosclerosis. 1983;3:23–30. doi: 10.1161/01.atv.3.1.23. [DOI] [PubMed] [Google Scholar]
- 43.Huang L, Sineva EV, Hargittai MRS, Sharma SD, Suthar M, Raney KD, Cameron CE. Protein Expression Purif. 2004;37:144–153. doi: 10.1016/j.pep.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 44.Blight KJ, McKeating JA, Rice CM. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.