Abstract
In mammalian RNA polymerase I transcription, SL1, an assembly of TBP and associated factors (TAFs), is essential for preinitiation complex formation at ribosomal RNA gene promoters in vitro. We provide evidence for a novel component of SL1, TAFI41 (MGC5306), which functions in Pol I transcription. TAFI41 resides at the rDNA promoter in the nucleolus and co-purifies and co-immunoprecipitates with SL1. TAFI41 immunodepletion from nuclear extracts dramatically reduces Pol I transcription; addition of SL1 restores the ability of these extracts to support Pol I transcription. In cells, siRNA-mediated decreased expression of TAFI41 leads to loss of SL1 from the rDNA promoter in vivo, with concomitant loss of Pol I from the rDNA and reduced synthesis of the pre-rRNA. Extracts from these cells support reduced levels of Pol I transcription; addition of SL1 to the extracts raises the level of Pol I transcription. These data suggest that TAFI41 is integral to transcriptionally active SL1 and imply a role for SL1, including the TAFI41 subunit, in Pol I recruitment and, therefore, preinitiation complex formation in vivo.
Keywords: ribosomal RNA synthesis, RNA polymerase I, RNA polymerase recruitment, SL1, TAFI41
Introduction
Accurate initiation of transcription by the three eukaryotic RNA polymerases, with few exceptions, requires a complex of TBP (TATA-box-binding protein) and TBP-associated factors (TAFs), which perform distinct roles in mediating a specific interaction between the polymerases and their target promoters (Hernandez, 1993; Goodrich and Tjian, 1994; Hahn, 1998). The complement of TAFs in each complex is distinct to each class (Pol I, II or III) of genes and their cognate promoters.
There are substantial similarities in Pol I transcription of rDNA genes between organisms as diverse as yeast and mammals (reviewed in: Hannan et al, 1998; Grummt, 1999; Reeder, 1999; Nomura, 2001; Moss and Stefanovsky, 2002; Grummt, 2003; Moss, 2004; Russell and Zomerdijk, 2005). In mammalian Pol I transcription in vitro, TBP–TAF complex SL1 (selectivity factor 1), known as TIF-IB in mouse, is essential (Comai et al, 1992, 1994; Zomerdijk et al, 1994; Heix et al, 1997; Friedrich et al, 2005) and upstream binding factor UBF functions as an activator of transcription (Bell et al, 1988; Jantzen et al, 1990; Hisatake et al, 1991; O'Mahony and Rothblum, 1991; Jantzen et al, 1992; Kuhn and Grummt, 1992). UBF can influence rRNA gene expression at multiple levels, functioning as an activator at promoter escape by Pol I in vitro (Panov et al, 2006), as a regulator (repressor) of elongation (Stefanovsky et al, 2006) and as an antirepressor (Kuhn and Grummt, 1992; Brou et al, 1993; Pelletier et al, 2000) in the context of chromatin, and has been implicated in large-scale chromatin decondensation in vivo (Chen et al, 2004; Mais et al, 2005).
The human SL1 complex of ∼300 kDa includes TBP and the three TAFIs TAFI48, TAFI63 and TAFI110 (Comai et al, 1992, 1994; Zomerdijk et al, 1994). SL1 from human and rodents can bind the promoter independently and direct specific initiation of transcription in the absence of UBF (Smith et al, 1990, 1993; Schnapp and Grummt, 1991; Friedrich et al, 2005). Core promoter binding is mediated by the TAFIs in SL1 (Rudloff et al, 1994; Beckmann et al, 1995). Another important role for the TAFIs in SL1 inferred from in vitro analyses is in the recruitment of a functional Pol I complex to the start site via its interaction with human RRN3 (murine TIF-IA), a component of initiation-competent Pol Iβ (Miller et al, 2001; Yuan et al, 2002; Friedrich et al, 2005). Furthermore, at least two subunits of SL1 interact with UBF (Kwon and Green, 1994; Beckmann et al, 1995), and these interactions prevent the otherwise rapid dissociation of UBF from the rDNA promoter (Friedrich et al, 2005). Taken together, the in vitro data imply that SL1 drives or nucleates preinitiation complex (PIC) formation at the rDNA promoter, leading to productive initiation of transcription by Pol I. Reconstitution of transcription in vitro using recombinant SL1 comprised of the four previously characterized components has been difficult and inefficient, suggesting that additional components and/or specific modifications might be required to assemble a fully active SL1 (Heix et al, 1997). Here, we describe our identification of a novel TAF component of SL1 (TAFI41) and present evidence that TAFI41 plays an important role in Pol I transcription. In the course of these studies, we also uncovered evidence to suggest that SL1, including the TAFI41 subunit, is required for Pol I recruitment and, thus, PIC formation in Pol I transcription in vivo.
Results and discussion
Human nucleolar protein TAFI41 co-purifies with the SL1 protein complex and activity
Silver staining of TBP-antibody affinity purified SL1 revealed only four major protein bands corresponding to TBP, and TAFIs 48, 63 and 110 (Comai et al, 1992). However, amino-acid sequence analyses of tryptic peptides derived from the TBP and TAFI48 region of the gel (Comai et al, 1994) revealed a peptide (KQMNVGEDLENEDFD) with no homology to any of the previously identified TAFs or TBP, matching amino-acid sequences within several human ESTs and encoded by a gene (JOSD3 (Josephine domain containing protein 3) or MGC5306; accession number NM_024116) located on human chromosome 11q21. Other peptide sequences with homology to the protein encoded by this gene (KAIFERFK and KLAGDSFIVSSEFPVRLSVY) were derived from the same region of the gel, mixed with peptide sequences from TAFI48 (KTSANISALIK and KGPVTDDEEV, respectively), confirming the presence of this protein in TBP-antibody affinity purified SL1. The encoded protein of 278 amino acids (Supplementary Figure S1) has a calculated molecular weight of 32 kDa, but an apparent molecular weight of ∼41 kDa. (Intriguingly, a faint band of ∼41 kDa is sometimes visible on protein gels following silver staining of TBP-antibody affinity purified SL1 (Supplementary Figure S2A).) This protein has been reported to interact in a yeast two-hybrid assay with DNA polymerase β and a possible function in base excision repair was therefore suggested (Wang et al, 2004). We provide evidence that this protein is also an SL1-specific TBP-associated factor (TAFI) involved in Pol I transcription and refer to it hereafter as TAFI41.
TAFI41 contains a basic stretch of amino acids including a putative NLS (83–108), a serine-rich region in the N-terminal quarter of TAFI41 and two acidic regions (184–192 and 200–216), and these features are conserved in a putative mouse homologue, mTAFI41 (accession number: NM_029248), of 322 amino acids and a calculated molecular weight of 37 kDa (Supplementary Figure S1) (variants NM_026541 and NM_027261 of 288 and 290 amino acids, respectively, and calculated molecular weight 33 kDa, are truncated in the C-terminus). mTAFI41 shows 56% identity and 63% similarity to human TAFI41 in the first 278 amino acids (Supplementary Figure S1).
To explore the association between TAFI41 and human SL1 from cells, SL1 was extensively purified from HeLa nuclear extracts as described in Figure 1A and B. Fractions from salt-gradient elution of the POROS Heparin column were analysed for the presence of TAFI41 by immunoblotting, using an antibody generated against three TAFI41 peptides (TAFI41-peptides antibody), and for the ability to support SL1-dependent rDNA promoter-specific transcription. TAFI41 co-purified with TAFI110 and TAFI63 subunits of SL1 and with SL1 activity (Figure 1B; POROS Heparin). The desalted 0.7 M POROS Heparin fraction of SL1 was applied to an SP-Sepharose column, step-eluted and applied to a TBP-antibody affinity column, yielding extensively purified SL1 (Supplementary Figure S2A, shows a silver-stain gel of SL1 similarly purified by Heparin Agarose, SP-Sepharose and TBP-antibody affinity chromatography). TAFI41 co-purified with SL1 eluted from the TBP antibody column (Figure 1B, TBP-antibody affinity column, lane 5). The peptide-eluted SL1 was passed over a Mono S column. Significantly, this extensively purified SL1 preparation demonstrated TAFI41 in the fractions that contained SL1 activity (Figure 1B; Mono S).
Figure 1.
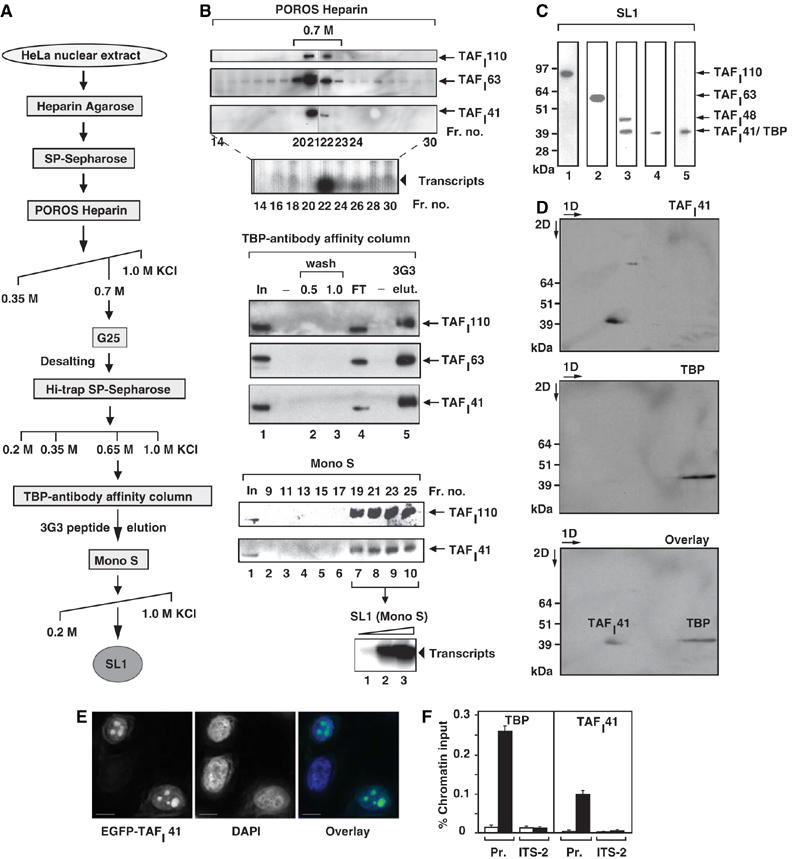
Human nucleolar protein TAFI41 co-purifies with TBP-antibody affinity-purified SL1 and is found at the rDNA promoter in human cells. (A) Human SL1 purification scheme. For analytical purification of SL1, HeLa cell nuclear extract was applied to POROS Heparin Agarose and SP-Sepharose columns, as previously described (Comai et al, 1992; Friedrich et al, 2005), then salt gradient eluted from a POROS Heparin column. The SL1 fractions were pooled, desalted on G25 Sepharose, step-eluted from SP-Sepharose and applied to a TBP-antibody affinity column. SL1 complexes were eluted with an excess of TBP (3G3) peptide and this eluate was applied to a Mono S column, which was developed by a linear salt gradient (KCl at concentrations (M) as indicated). (B) TAFI41 co-purifies with SL1 protein and activity from HeLa cell nuclear extracts. Fractions obtained by salt gradient elution from the POROS Heparin column (B) were analysed for SL1 protein by immunoblotting, with antibodies specific for SL1 subunits TAFI110 and TAFI63, and an antibody generated against three peptides of TAFI41 (see Materials and methods). The bulk of TAFI41 eluted with the other SL1 subunits at 0.7 M KCl (fractions 21 and 22). Fraction 22 supported specific SL1-dependent reconstituted Pol I transcription. The 0.7 M KCl fraction was desalted to 0.2 M KCl, passed over SP-Sepharose and a salt-step elution was performed. The bulk of SL1 including TAFI41 eluted at 0.65 M KCl and supported specific SL1-dependent transcription. This SL1 fraction was loaded onto a TBP-antibody affinity column (input, lane 1), some of which flowed through (FT, lane 4). The column was washed at 0.5 and 1.0 M KCl (lanes 2 and 3). Bound proteins from the washed column were eluted with excess TBP-epitope peptide 3G3 (1 mg/ml) (lane 5) and fractions were immunoblotted for SL1 with antibodies specific for TAFI110 and TAFI63, and the TAFI41-peptides antibody. SL1 was concentrated and purified away from the 3G3-peptide on a Mono S column, developed with a linear salt gradient. The Mono-S purified SL1 fractions 19–25 (fractions 19, 21, 23 and 25 are shown in lanes 7–10) were pooled and tested for the ability to support SL1-dependent transcription with UBF and Pol I (lanes 1–3, arrowhead). (C) TBP and TAFI41 display similar apparent molecular weights. SL1 (the 0.7 M fraction from the POROS-Heparin column, see A, B) was immunoblotted with antibodies specific for TAFI110, TAFI63, TAFI48 and TBP (top panel, lanes 1, 2, 3 and 5, respectively) and the TAFI41-peptides antibody (lane 4). A band of similar mobility to those of TBP and TAFI41, detected by TAFI48 antibodies, is either a degradation product of TAFI48 or could represent a protein encoded by a splice variant 2 of TAFI48 (top panel, lane 3). (D) TBP and TAFI41 are separable by two-dimensional gel electrophoresis. SL1 (the 0.7 M fraction from the POROS-Heparin column, see A and B) was subject to first dimension isoelectric focusing (pH 3–10), then to second dimension separation (4–12% Bis–Tris gradient gel, Invitrogen). SL1 proteins were immunoblotted, probed first with TAFI41-peptides antibody (upper panel) and then with antibodies specific for TBP (middle panel). The lower panel shows an overlay of the signals from these immunoblots. (E) Overexpressed TAFI41 accumulates in nucleoli. HeLa cells were transfected with expression vectors pEGFP-TAFI41. After 24 h, the cells were fixed and viewed by confocal microscopy. DNA was stained with DAPI. Overlay is a composite of the DAPI (blue) and EGFP-TAFI41 (green) signals. Scale bar, 10 nm. (F) ChIP analysis indicates that TAFI41 is present at the promoter of the rRNA genes. ChIP analysis (from HeLa cells) used TAFI41-peptides antibody (and as a control rabbit preimmune serum) or a TBP mouse monoclonal antibody (and as a control mouse IgG), followed by quantitative real-time PCR with primers specific for the promoter region and the internal transcribed spacer (ITS-2) of the human rDNA (see Materials and methods). The relative levels of rDNA associated with TAFI41 and TBP at these sites are expressed as percentage of input chromatin and are from two independent experiments.
Immunoblotting with TAFI41-peptides antibody demonstrated that TAFI41 co-migrates with TBP at ∼40 kDa (Figure 1C, compare lanes 4 and 5); separation was achieved in 2D gels owing to the difference in the pIs of these proteins (Figure 1D). The electrophoretic mobility of TAFI41 shifted upon dephosphorylation (data not shown), indicating that TAFI41 is a phosphoprotein. Collectively, these data provide evidence to suggest that TAFI41 is a previously overlooked subunit of human SL1. The comigration of TAFI41 with TBP may explain why it had remained elusive for more than 10 years following the initial identification of SL1 subunits (Comai et al, 1994; Zomerdijk et al, 1994). Consistent with a role for TAFI41 as a component of SL1 in Pol I transcription, overexpressed EGFP-TAFI41 accumulated in the nucleoli, the site of Pol I transcription, (Figure 1E) and chromatin immunoprecipitation (ChIP) assays detected TAFI41 and TBP at the rDNA promoter in cells (Figure 1F).
TAFI41 co-immunoprecipitates with SL1 from human cells
Co-precipitation of EGFP-TAFI41 with the endogenous SL1 subunits TAFI110, TAFI63 and TBP (Figure 2A) was detected following immunoprecipitation, using GFP-specific antibodies, from human cells expressing EGFP-TAFI41. In a similar experiment with Flag-tagged TAFI41, we also demonstrated co-precipitation of TAFI41 with the SL1-subunits (Figure 2B, lane 7). Conversely, endogenous TAFI41 co-precipitated with Flag-TAFI63 following immunoprecipitation, using Flag-specific antibodies, from human cells expressing Flag-tagged TAFI63 (Figure 2B, lane 5).
Figure 2.
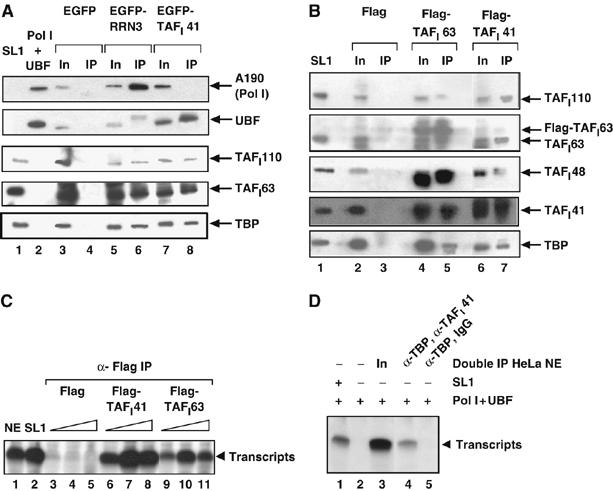
TAFI41 is a component of transcriptionally active SL1. (A) EGFP-TAFI41 co-immunoprecipitates with SL1 and UBF from human cells. Immunoprecipitation (IP) with GFP-specific antibodies was from HEK293 whole-cell extracts 48 h post-transfection of the cells with pEGFP-TAFI41 (lane 8), empty expression vector (lane 4) or pEGFP-RRN3 (lane 6). Immunoblotting used antibodies specific for the Pol I largest subunit A190, UBF and SL1 subunits TAFI110, TAFI63 and TBP. Input (In), 10% of extract used for IP (lanes 3, 5 and 7). SL1 (lane 1), and Pol I and UBF (lane 2) were loaded as markers. (B) Endogenous TAFI41 and Flag-TAFI41 co-immunoprecipitate with SL1 from human cells. IP with Flag-specific antibodies was from HEK293 whole-cell extracts 48 h post-transfection of the cells with pcDNA-Flag-TAFI41 (lane 7), empty expression vector (lane 3) or pcDNA-Flag-TAFI63 (lane 5). Immunoblotting used antibodies specific for SL1 subunits TAFI110, TAFI63, TAFI48, TBP and TAFI41-peptides antibody. Input (In), 80% of extract used for IP (lanes 2, 4 and 6). Purified SL1 (lane 1) was loaded as a marker. (C) TAFI41-associated proteins from cells support SL1-dependent Pol I transcription. Immunoprecipitated complexes of 40 (lanes 3, 6 and 9), 80 (lanes 4, 7 and 10) and 160 ng (lanes 5, 8 and 11) prepared in B, from HEK293 cells transfected with pcDNA-Flag-TAFI41 (lanes 6–8), empty vector (lanes 3–5) or pcDNA-Flag-TAFI63 (lanes 9–11), were tested for SL1-dependent activity in a specific transcription assay with Pol I, UBF and the rDNA promoter. Controls are transcription in a HeLa nuclear extract (lane 1) and from a partially purified SL1 fraction (the 0.7 M fraction from the POROS-Heparin column, see A, B; lane 2) added to the reconstituted transcription assay. (D) Endogenous TAFI41 co-immunoprecipitates with TBP-containing complexes from human cells and the TAFI41-TBP-containing complexes support rDNA promoter-specific Pol I transcription. TBP-containing complexes immunoprecipitated from HeLa cell nuclear extracts using TBP-specific monoclonal antibody (3G3) beads were eluted from the beads using an excess of epitope peptide, then incubated with TAFI41-peptides antibody in a second IP. The SL1-dependent transcription activity of the TBP-TAFI41-containing complexes was determined in an in vitro transcription assay with Pol I and UBF (lane 4). In the control sample, IgG was used in the second immunoprecipitation reaction in place of the TAFI41-peptides antibody (lane 5). The SL1 activity of the input Hela nuclear extracts is shown in lane 3. Lanes 1 and 2 contain control reconstituted transcription reactions with Pol I and UBF, with and without SL1, respectively.
Furthermore, intact and functional SL1 was immunoprecipitated, with Flag-specific antibodies, from human cells expressing Flag-TAFI41; the (Flag-peptide-eluted) immunocomplexes were capable of directing SL1-dependent accurate transcription initiation in reactions reconstituted with Pol Iβ and the rDNA promoter (Figure 2C, lanes 6, 7 and 8). SL1-dependent transcription was also observed with Flag immunocomplexes from extracts of cells expressing Flag-TAFI63, a previously established subunit of SL1 (the difference in transcription levels perhaps reflecting differences in the specific activities and/or yields of functional SL1 complexes incorporating Flag-TAFI41 or Flag-TAFI63). Moreover, SL1 activity was detectable in immunocomplexes precipitated with TBP-specific antibodies, eluted with TBP-epitope peptide, and then re-precipitated with TAFI41-specific antibody but not with IgG antibodies (Figure 2D, lanes 4 and 5, respectively). Therefore, TAFI41 appears to be an integral component of at least some portion of transcriptionally active SL1 complexes.
SL1 components TAFI48, TAFI63 and TAFI110 have previously been shown to interact with each other and with TBP in GST pull-down experiments and in binding assays with in vitro translated or baculovirus-Sf9 cell-expressed proteins (Comai et al, 1994). TAFI41 interacts detectably with the other TAFs of SL1 and, weakly, with TBP and RRN3 in such assays and with TAFI110 in Far Western analyses (see Supplementary Figure S3). Although it is not known whether such interactions occur in the context of the SL1 complex or indeed the PIC, such interactions would be consistent with TAFI41 being a subunit of the stable multiprotein complex SL1, with a potential role in the assembly of productive Pol I PICs.
The majority of the transcriptionally active SL1 complexes contain TAFI41
We next asked whether immunodepletion of TAFI41 from nuclear extracts would deplete a significant amount of SL1 activity. A TAFI41 antibody generated against amino acids 98–204 of TAFI41 (antibody D) selectively immunoprecipitated SL1 subunits TAFI110, TBP and TAFI41 from HeLa nuclear extract (Figure 3A, lane 2 compared to lane 4) and, therefore, partially immunodepleted these SL1 subunits from the extract (the partial immunodepletion of TAFI41 and TBP is detectable in Figure 3A, lane 1 compared to lane 3). The TAFI41 antibody D-depleted extract supported a reduced level of Pol I transcription (Figure 3B, lane 1 compared to lane 3). Addition of SL1 to the TAFI41 antibody D-depleted extract restored the ability of the extract to support Pol I transcription (Figure 3B, lane 2). These results suggest that most transcriptionally active SL1 complexes contain TAFI41.
Figure 3.
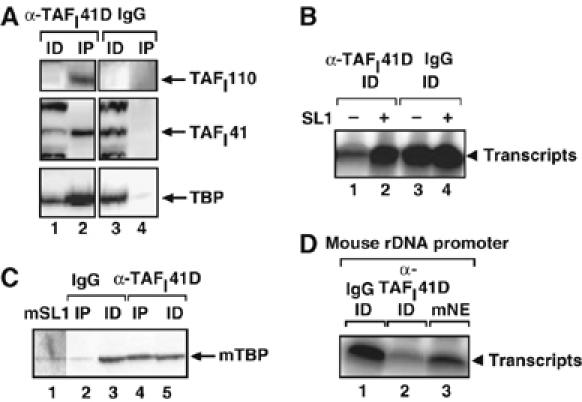
Human TAFI41 antibodies deplete SL1 activity from human and mouse nuclear extracts. (A) Human TAFI41 antibodies deplete SL1 subunits from HeLa cell nuclear extract. HeLa nuclear extracts were incubated with hTAFI41 antibody D (raised against a recombinant human TAFI41 deletion mutant, see Materials and methods; lanes 1 and 2) or control (IgG; lanes 3 and 4) antibodies overnight at 4°C. Immunoprecipitates (IP, lanes 2 and 4) and 10% of the immunodepleted extracts (ID, lanes 1 and 3) were immunoblotted with antibodies specific for TAFI110, TBP and TAFI41-peptides antibody. The signal in the ID for TAFI110 (lanes 1 and 3) was too high and this resulted in a ‘white-out' following chemiluminescence. (B) Human TAFI41 antibodies deplete SL1 transcriptional activity from HeLa cell nuclear extract. Extracts from (A), immunodepleted (ID) using hTAFI41 antibody D (lanes 1 and 2) or control (IgG, lanes 3 and 4), were analysed in a specific transcription assay with the human rDNA promoter, in the absence (lanes 1 and 3) or presence of added purified SL1 (Superose 6 fraction; 1 μl, lanes 2 and 4). Transcripts were analysed by S1 nuclease protection. (C) Human TAFI41 antibodies immunoprecipitate mouse TBP from mouse 3T3 nuclear extract. Mouse 3T3 nuclear extract (25 μl) was incubated with hTAFI41 antibody D (lanes 4 and 5) or control (IgG, lanes 2 and 3) overnight at 4°C. Immunoprecipitated complex (IP) and 10% of the immunodepleted extract (ID) were immunoblotted with antibodies specific for TBP. Partially purified mouse SL1 (TIF-IB) (lane 1) was loaded as a marker. (D) Human TAFI41 antibodies partially deplete Pol I transcription activity from mouse cell nuclear extract. In all, 15% of the extracts immunodepleted by hTAFI41 antibody D (lane 2) or control (lane 1) antibodies were analysed in a specific transcription assay using the mouse rDNA promoter. Transcripts were analysed by S1 nuclease protection with a 5′-end labelled oligonucleotide complementary to the first 40 nt of the mouse pre-rRNA. Lane 3 contains the control transcription reaction with mouse nuclear extract (mNE).
A mouse TAFI41 orthologue in mSL1 (TIF-IB)
The discovery of a potential mouse orthologue in the sequence database prompted us to try and determine whether a functional TAFI41 exists in mouse cells. There are no bands detectable at or around the calculated molecular weight of 37 kDa for the mouse homologue in TBP-affinity purified mouse SL1 (TIF-IB) (Rudloff et al, 1994), although reduced silver staining and/or co-migration with, for example, mTAFI48 could not be excluded. The human TAFI41-peptides antibody did not detect a potential mouse homologue following immunoblotting of mouse nuclear extracts (data not shown). Nonetheless, human TAFI41 antibody D immunoprecipitated TBP from mouse nuclear extracts (Figure 3C, lane 4 compared to 2) and reduced Pol I transcription in these extracts (Figure 3D, lane 2 compared to 1), suggesting that mouse TAFI41 might be a component of transcriptionally active mSL1 (TIF-IB) complexes. SL1 is a major determinant in the species specificity of Pol I transcription (Heix and Grummt, 1995). Overexpression of human TAFI41 in mouse cells did not reprogramme these cells to transcribe a human rDNA promoter (data not shown).
siRNA-mediated downregulation of TAFI41 expression in cells decreases Pol I transcription
A positive role for TAFI41 in Pol I transcription is suggested by the findings that TAFI41 is a component of active SL1 complexes. To determine whether or not TAFI41 is required for Pol I transcription in cells, we downregulated its expression using siRNAs. Human embryonic kidney 293 (HEK293) cells transfected with TAFI41-siRNA showed reduced levels of TAFI41 mRNA (data not shown) and protein (to less than 25%; Figure 4A, lanes 1–3 compared to lanes 4–6). Concomitant with downregulated TAFI41 expression, a decreased level of 47S pre-rRNA was detected in these cells (Figure 4B, lanes 1–3 compared to lanes 4–6, and bar graph). TAFI41-siRNA also mediated downregulation of TAFI41 expression (Figure 4C) and decreased pre-rRNA levels (Figure 4D, lane 1) in SJSA cells, in contrast to the control siRNA (Figure 4C and D, lane 4). siRNA-mediated downregulation of the expression of SL1 subunit TAFI48 or essential Pol I-associated factor RRN3 (Figure 4C) similarly reduced Pol I transcription (Figure 4D, lanes 2 and 3, respectively). Consistent with these findings, nuclear extracts from TAFI41-siRNA-treated cells supported a reduced level of Pol I transcription (Figure 4E, lane 1 compared to 4) and an increased level of transcription was detectable in these extracts following addition of SL1 (Figure 4E, lanes 2 and 3). Taken together, these results suggest that siRNA-mediated downregulation of TAFI41 expression in cells specifically impairs the function of SL1, implying a role for TAFI41 in SL1 function.
Figure 4.
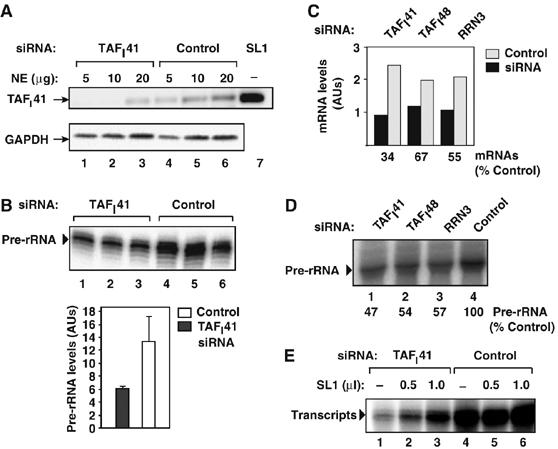
siRNA-mediated downregulation of TAFI41 expression in cells leads to a decrease in Pol I transcription levels. (A) TAFI41-specific siRNA reduces TAFI41 protein levels in cells. Nuclear extracts (5, 10 and 20 μg, respectively) of HEK293 cells transfected with TAFI41-specific siRNA (lanes 1–3) or scrambled siRNA (control; lanes 4–6) were immunoblotted using TAFI41-peptides antibody and GAPDH-specific antibody. Lane 7 contains SL1 as a marker. (B) TAFI41-specific siRNA reduces Pol I transcription levels in cells. Pre-rRNA levels in total RNA extracted from HEK293 cells transfected, in triplicate, with TAFI41-specific siRNA (lanes 1–3) or scrambled siRNA (control; lanes 4–6) were analysed by S1 nuclease protection with an oligonucleotide complementary to the first 40 nt of the 47 S pre-rRNA. Combined data are represented in the graph. (C) siRNA-mediated reduction of hTAFI41 TAFI48 and RRN3 mRNA levels. Messenger RNA levels in total RNA from SJSA cells transfected with TAFI41-, TAFI48- and RRN3-specific or scramble (control) siRNAs were determined by RT–PCR and normalized to β-actin mRNA levels. (D) siRNA-mediated reduction of TAFI41, TAFI48 or RRN3 gene expression reduces pre-rRNA synthesis in cells. Pre-rRNA levels in total RNA extracted from the siRNA-treated SJSA cells (see C) were analysed by S1 nuclease protection as in (B). (E) Pol I transcription in extracts of TAFI41-specific siRNA-treated cells can be partially restored by addition of SL1. Nuclear extracts (30 μg) of HEK293 cells transfected with siRNAs specific for TAFI41 (lanes 1–3), or scrambled (control; lanes 4–6) were analysed for specific transcription activity in the absence (lanes 1 and 4) or presence of purified SL1 (Superose 6 fraction; 0.5 μl, lanes 2 and 5, or 1 μl, lanes 3 and 6).
Downregulation of TAFI41 expression decreases the rDNA promoter occupancy of SL1 and Pol I in cells
The observed reductions in Pol I transcription in cells transfected with siRNAs for the TAFI48 and TAFI41 subunits of SL1 (Figure 4) support an in vivo role for SL1 in Pol I transcription of the rRNA genes. To provide further evidence for this and examine the role of SL1 and TAFI41 in Pol I transcription in vivo, we downregulated TAFI41 expression in cells and analysed the consequences for SL1 and Pol I occupancy at the rDNA repeat by ChIP. siRNA-mediated downregulation of TAFI41 expression not only impaired SL1 function (Figure 4) but also drastically decreased the rDNA promoter occupancy of SL1 subunit TAFI110 and Pol I subunit A135 (Figure 5). SL1 can direct PIC formation in vitro (Smith et al, 1990, 1993; Schnapp and Grummt, 1991; Friedrich et al, 2005), but a role for SL1 in this process in vivo had not been demonstrated. Our finding that the presence of Pol I at the rDNA promoter is dependent on the presence of SL1 provides evidence to suggest that SL1, including TAFI41, is required for Pol I recruitment and, hence, PIC formation in vivo. This is consistent with the observed dramatic decrease in Pol I A135 occupancy in transcribed regions of the rDNA repeat (Figure 5).
Figure 5.
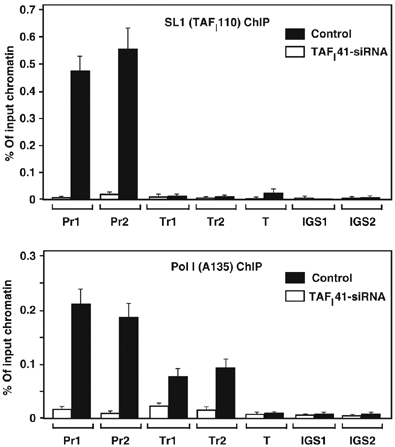
Reduced occupancy of SL1 and Pol I at the rDNA in TAFI41-siRNA-treated cells. ChIP analysis of control or TAFI41-siRNA-treated HEK293 cells (72 h post-transfection) with antibodies specific for TAFI110 (SL1) or A135 (second largest subunit of Pol I) and with an array of primers sets for the human rDNA repeat (Pr1 and Pr2: UCE and core promoter sequences; Tr1 and Tr2, the transcribed 18S and 28S rRNA gene sequences; T, sequences downstream of terminator; IGS1 and IGS1, sequences in pseudo-gene CDC27 and p53-binding moiety in the intergenic spacer of the rDNA repeat; see Materials and methods). The levels of rDNA associated with TAFI110 (SL1) and A135 (Pol I) at each site (relative to control antibodies) are expressed as percentage of input chromatin.
TAFI41 interacts with UBF and might function as a coactivator in UBF-activated transcription
TAFI41 overexpressed in cells co-immunoprecipitated with UBF from cell extracts (Figure 2A, lane 8). Interaction between TAFI41 and UBF was observed in affinity chromatography with Ni-NTA-immobilized His-TAFI41 (from recombinant baculovirus-infected insect cells) using in vitro translated UBF (Figure 6A, lane 2) and in GST-TAFI41 pull downs of baculovirus-Sf9 cell-expressed recombinant UBF (Figure 6B, lane 2). Moreover, Far-western analysis also demonstrated that in vitro translated 35S-labelled TAFI41 interacted with renatured recombinant UBF (Figure 6C, lane 2).
Figure 6.
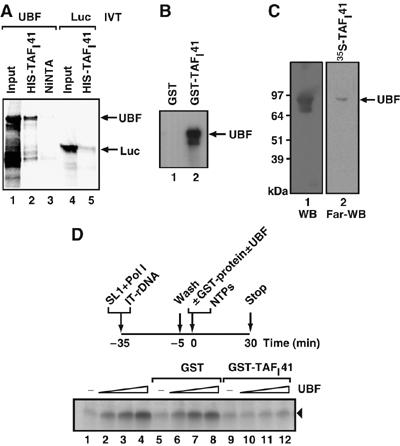
TAFI41 interacts with UBF and can squelch UBF-dependent activation of Pol I transcription. (A) Recombinant His-TAFI41 interacts directly with in vitro translated UBF. His-TAFI41 (lanes 2 and 5), baculovirus-expressed in insect cells and purified on NiNTA beads, was incubated with in vitro translated (IVT) 35S-radiolabelled UBF (lanes 1–3) or luciferase (negative control protein; lanes 4 and 5). Empty NiNTA beads were included as a control (lane 3). Beads were washed and bound proteins analysed by autoradiography. (B) GST-TAFI41 interacts directly with recombinant UBF. GST-TAFI41 (lane 2) or GST (lane 1) proteins on beads were incubated with purified recombinant baculovirus-Sf9 cell-expressed human UBF1. Beads were washed and bound proteins analysed by immunoblotting with UBF-specific antibodies. (C) A direct interaction between TAFI41 and UBF is detectable by Far-Western analysis. In vitro translated 35S-radiolabelled TAFI41 was incubated with membrane-bound, then denatured and renatured UBF protein (Far-WB; lane 2). Binding was analysed by autoradiography. In control lane 1, denatured–renatured UBF was detected by UBF-specific antibodies following Western blotting (WB). (D) TAFI41 squelches UBF-activated but not basal SL1-directed Pol I transcription. Preinitiation complexes (PICs) of SL1 (Superose 6 fraction) and Pol I were assembled on an immobilized human rDNA promoter template (IT-rDNA, Fr4; Panov et al, 2001) for 30 min on ice. The PICs were washed in TM10/0.075 M KCl, then GST-TAFI41 (50 ng) was added concomitantly with 0, 1, 10 and 20 ng of UBF (lanes 9, 10, 11 and 12, respectively) and NTPs and transcription was analysed by S1 nuclease protection. In the control samples, in place of GST-TAFI41, no protein (lanes 1–4) or GST (50 ng; lanes 5–8) was used.
To analyse the functional significance of the UBF–TAFI41 interaction, we tested the effect of TAFI41 upon UBF-activated transcription in a reconstituted transcription reaction. Preinitiation complexes of SL1 and Pol I were assembled on an immobilized rDNA promoter template. Addition of recombinant TAFI41 did not affect basal transcription from these templates (Figure 6D, lane 9 compared to lanes 1 and 5). However, UBF-activated transcription was blocked by addition of recombinant TAFI41 (Figure 6D, lanes 10–12). This squelching of UBF activation is likely to be specific, because GST alone (Figure 6D, lanes 6–8) did not produce the effect. These data suggest a potential coactivator function for TAFI41, but we do not yet have direct evidence for this. We are currently working to improve the efficiency and efficacy of reconstitution of Pol I transcription using SL1 assembled from its individual subunits TBP and TAFIs, 110, 63, 48 and 41, to enable further study of the individual roles of TAFI41 and the other SL1 subunits.
In conclusion, we have demonstrated that TAFI41 is a novel component of transcriptionally active SL1, important in Pol I transcription. Interestingly, the introns of the TAFI41 gene (MGC5306) encode six H/ACA box snoRNAs and two C/D box snoRNAs (Kiss et al, 2004), providing a link between transcription of the 47S pre-rRNA by Pol I and its processing and modification to produce the 18S, 5.8S and 28S rRNAs. We have observed a correlation between TAFI41 expression levels and Pol I transcription activity in cells. Furthermore, our data implicate the SL1 complex, including TAFI41, in recruitment of the Pol I complex to the rDNA promoter in vivo, perhaps through an interaction with RRN3/TIF-IA. We speculate that the TAFI41 subunit of SL1 might also be required to upregulate Pol I transcription for sustained hyperproliferation of cells, particularly given the observed overexpression of TAFI41 (MGC5306) in certain tumours ((Wang et al, 2004); Pathak and Zomerdijk, unpublished results). The importance of TAFI41 with respect to cell fate is also suggested by the finding that decreased expression of TAFI41 activates the p53 response pathway (Pathak and Zomerdijk, unpublished results), induces apoptosis and reduces cell proliferation (Wang et al, 2004) most likely as a consequence of the downregulation of Pol I transcription.
Materials and methods
TAFI41 antibodies
TAF141 peptides (MDKSGIDSLDH; ENEKNAPWRKIL; KAKNTGQRGLKM), coupled to KLH, were used to generate rabbit polyclonal antibodies. This antibody is referred to as TAFI41-peptides antibody and is used throughout this study primarily for probing immunoblots. The characterization of the TAFI41-peptides antibody specificity is described (Supplementary Figure S2B and C).
Human TAFI41 protein D (amino acids 98–204) was cloned in pGEX-4T vector and expressed in Escherichia coli BL21(DE3). GST-TAFI41 (98–204) protein was affinity purified on glutathione–Sepharose and after thrombin cleavage to release the TAFI41 protein portion from GST, purified TAFI41 protein was used to generate polyclonal sheep antibodies (Scottish National Blood Transfusion Service), which were affinity purified on HiTRAP NHS-activated HP coupled to purified hTAFI41 protein D according to the manufacturer's instructions (Amersham Biosciences). This antibody was used in the immunoprecipitation and depletion experiments of Figure 3.
Pol I transcription machinery components
Human SL1 was purified from HeLa cell nuclear extracts for analytical purposes as described in Figure 1A and B (and based in part on protocols described by Friedrich et al, 2005). For preparative scale purification, we purified SL1 through the following columns: Heparin Agarose, SP-Sepharose, POROS-Heparin, Mono S and Superose 6, as detailed previously (Miller et al, 2001; Friedrich et al, 2005); this purified SL1 is referred to as Superose 6 fraction SL1. UBF and Pol I were purified as described previously (Miller et al, 2001; Panov et al, 2006). Hela cell nuclei were purchased from the National Cell Culture Center (Minneapolis, MN, USA).
Immunoblotting and immunoprecipitation
Immunoblotting and immunoprecipitations used antibodies for human TAFI41 (TAFI41-peptides antibody, used in Westerns and IPs or TAFI41-antibody D, used in IPs; see above), TAFI110, TAFI63, TAFI48, RRN3, TBP (Comai et al, 1994; Zomerdijk et al, 1994; Heix et al, 1997; Miller et al, 2001), GAPDH (clone 6C5, Research Diagnostics Inc.), Flag (Sigma) and GFP (Roche). For immunoprecipitation of Flag- or EGFP-tagged proteins, HeLa cells were transfected (Effectene, Qiagen) with pcDNA-Flag-TAFI41, pcDNA-Flag-TAFI63 (Flag peptide coding sequence and full-length cDNA of human TAFI41 or TAFI63 subcloned into pcDNA3.2/V5-DEST vector, Invitrogen) or pEGFP-TAFI41, pEGFP-RRN3 expression vectors (full-length cDNA of human TAFI41 or RRN3 subcloned into pEGFP, Invitrogen), and ‘empty' expression vectors were used as controls in these transfections. Precleared nuclear extract (0.4 mg) from these transfected cells was incubated with Flag-specific or GFP-specific antibody beads in TM10/0.2 M KCl (50 mM Tris–HCl pH 7.9, 12.5 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM sodium-metabisulphite, 1 mM DTT, 0.015% NP40) for 2 h at 4°C, beads were washed and Flag precipitants were eluted by Flag peptide as described (Panova et al, 2006). For immunoprecipitation of endogenous proteins, 0.4 mg of precleared nuclear extract was incubated with antibody on Protein A Dynabeads in TM10/0.2 M KCl overnight at 4°C; beads were washed extensively in TM10/0.2.
In vitro transcription
In vitro Pol I transcription assays with human rDNA promoter (Fr 4, −193 to +239) or mouse rDNA promoter (−182 to +23) fragments were performed as described (Miller et al, 2001; Panov et al, 2001). The SL1 used for add-back into in vitro transcription reactions (Figures 3 and 4) was purified over the following columns: Heparin Agarose, SP-Sepharose, POROS-Heparin, Mono S and Superose 6, as detailed previously (Miller et al, 2001; Friedrich et al, 2005). This SL1 (Superose 6 fraction) is free of UBF and Pol I.
siRNA-mediated repression of hTAFI41, TAFI48 and RRN3 expression
HeLa, HEK293 or human astrocytoma SJSA cells (cultured in DMEM with 10% FCS) were transfected (Oligofectamine, Invitrogen) with synthetic siRNA (100 nM; Dharmacon) for hTAFI41 (target sequence 5′-AAGTGATTCATCAAGTGACTC), TAFI48 (5′-AAGAGGTACTCACC AATTATG), RRN3 (5′-AAATATGCGTGCATTAGAGAA) or Control (5′-AACAGTCGCGTTTGCGACTGG). After 48 h, nuclear extracts were prepared, and analysed by immunoblotting (with GAPDH serving as a reference) and/or used in in vitro transcription reactions; total RNA was isolated from the siRNA-treated cells to assess pre-rRNA levels by S1 nuclease protection as described (James and Zomerdijk, 2004). Messenger RNA levels were analysed by quantitative real-time PCR as outlined. Three siRNAs for TAFI41 were tested; the TAFI41-siRNA that caused the most effective repression of TAFI41 gene expression in cells is used throughout this study.
mRNA extraction and quantification by real-time PCR
Total RNA from siRNA-treated cell cultures was isolated using the RNeasy mini-kit (Qiagen), including an on-column DNase I treatment step. RNA (200 ng) was subsequently reverse-transcribed into cDNA with Superscript II (Invitrogen) using random hexamers. Real-time PCR (Taqman, Applied Biosystems) was carried out with an ABI Prism 7900 sequence detector using the following protocol: 50°C for 2 min, 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 1 min. Primers used were: for TAFI48: forward (f) primer 5′-TGCTCAGTGGTGCAGGAATG and reverse (r) 5′-CCTCCAATGGC CACAGTTTC, and Taqman probe: 6-FAM-ATTTTCCTTGGCTTCAAACATACG-TAMRA; for RRN3: (f) primer 5′-AGAACCAGCTGTT AGATCCAGACATA, and (r) 5′-TCAAGTACATGATAGAAGAACGGAA TTC and Taqman probe 6-FAM-AGGATGACCAGATCATCAACTGGCT-TAMRA.
TAFI41 mRNA was first reverse transcribed and then the cDNA was amplified using SYBR Green PCR master mix (Applied Biosystems) and specific primers in an ABI Prism 7900 system (Applied Biosystems). The primers used were: (f) 5′-CATGTGACATCTGATGCTGTGGAAC and (r) 5′-CGTGAACACTTTCAGGTGTACGAAC. PCR conditions used were: 50°C for 2 min, 95°C for 10 min, and 50 cycles of 95°C for 15 s and 60°C for 1 min. Primers and Taqman probe for β-actin were purchased from Applied Biosystems. Relative quantification of gene expression was performed using the comparative threshold (CT) method. Changes in mRNA expression levels were calculated following normalization to β-actin mRNA levels.
In vitro binding assays
Interactions of TAFI41 with UBF or luciferase were analysed using recombinant baculovirus-Sf9 cell-expressed His-TAFI41 (purified on Ni-NTA beads according to the manufacturer's instructions (Qiagen)) and 35S-labeled in vitro translated (TNT-coupled reticulocyte lysate system (Promega)) UBF or luciferase in in vitro binding assays.
In an alternative protein–protein binding assay, GST or GST-TAFI41 (expressed in E. coli and purified on glutathione–Sepharose according to the manufacturer (Amersham Biosciences)) was mixed with baculovirus-Sf9 cell-expressed and purified UBF (Panov et al, 2006).
In vitro binding assays were performed in 75 mM KCl in TM10 buffer plus 0.015% NP-40 for 30 min at 0°C. After binding, beads were extensively washed with 0.1 M KCl in TM10 and bound protein were analysed by autoradiography.
Far-Western analyses involved SDS–PAGE of recombinant purified UBF, transfer to membrane, denaturation and renaturation of membrane-bound UBF, incubation of the membrane with 35S-radiolabelled in vitro translated TAFI41 (using subclone pcDNA3.2/V5-DEST(Invitrogen)-TAFI41) and analysis of bound proteins by autoradiography as described (Miller et al, 2001).
Nucleolar chromatin immunoprecipitations
Nucleolar chromatin immunoprecipitations (ChIPs) were performed essentially as described by O'Sullivan et al (2002) with some modifications. HeLa or HEK293 cells were cultured to approximately 70% confluency, and the medium was refreshed 1 h before crosslinking. Crosslinking was performed for 10 min at 37°C by the addition of freshly made paraformaldehyde to a final concentration of 0.25–1%. Cells were collected into cold PBS supplemented with 1 mM AEBSF, pelleted, washed with PBS, and resuspended in high-magnesium buffer (10 mM HEPES pH 7.5, 0.88 M sucrose, 12 mM MgCl2, 1 mM DTT, Complete protease inhibitors (Roche)). Nucleoli were released by sonication, pelleted, resuspended in low-magnesium buffer (10 mM HEPES pH 7.5, 0.88 M sucrose, 1 mM MgCl2, 1 mM DTT, Complete protease inhibitors (Roche)), further sonicated and pelleted. The nucleoli were then resuspended in 1% SDS, 10 mM EDTA (pH 8), 50 mM Tris–HCl (pH 8), Complete protease inhibitor, and further SDS was added to a final concentration of 2%. After the lysis of the nucleoli (37°C for 15 min), the solution was diluted five-fold with TE buffer containing Complete protease inhibitors, sonicated, the resulting sheared nucleolar chromatin was centrifuged (15 000 g for 1 min), and the supernatant was used in nucleolar ChIP assays after quantification of input total DNA and determining the size of resulting sheared nucleolar DNA (fragments in the 250–300 bp size range).
Nucleolar ChIPs were performed using TBP-antibody (mouse monoclonal 3G3, kind gift from Dr L Tora), TAFI41-peptides rabbit polyclonal antibody, TAFI110 rabbit polyclonal antibody (Zomerdijk et al, 1994), A135 (Pol I second largest subunit) goat polyclonal antibody (Santa Cruz Biotech, Inc.) or the appropriate control IgG (Sigma) or preimmune serum immobilized on Protein G para-magnetic beads (Dynal). QPCR analysis was performed using SYBR Green PCR Master Mix and ABI7000 (both Applied Biosystems). For the ChIP analysis in Figure 1, we used the following primer pairs in the human rDNA repeat (positions according to GenBank Acc No U13369): 42 918–42 936 and 7–33 (core promoter); 7064–7090 and 7266–7291 (ITS-2). For ChIP analysis in Figure 6, the following primer pairs were used: 42 787–42 829 and 42 832–42 881 (Pr1: promoter UCE); 42 896–42 940 and 42 949–42 993 (Pr2: core promoter); 4501–4545 and 4656–4700 (Tr1: 18S rRNA gene); 12 100–12 144 and 12 260–12 304 (Tr2: 28S rRNA gene); 13 487–13 531 and 13 680–13 724 (just downstream of terminator, T); 35 822–35 866 and 35 987–36 031 (intergenic spacer IGS1: CDC27 pseudo-gene); 39 325–39 369 and 39 478–39 522 (IGS2: p53-binding moiety).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Information
Acknowledgments
We thank Lucio Comai, Arie Admon and Robert Tjian for analysis and amino-acid sequencing of components of SL1 and Laura Trinkle-Mulcahy for help with the fluorescence microscopy. We thank Robert Tjian and our colleagues in the Zomerdijk laboratory for advice and critical reading of the manuscript. JJG received a BBSRC PhD-studentship. JCBMZ is a Wellcome Trust Senior Research Fellow in the Basic Biomedical Sciences.
References
- Beckmann H, Chen JL, O'Brien T, Tjian R (1995) Coactivator and promoter-selective properties of RNA polymerase I TAFs. Science 270: 1506–1509 [DOI] [PubMed] [Google Scholar]
- Bell SP, Learned RM, Jantzen HM, Tjian R (1988) Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 241: 1192–1197 [DOI] [PubMed] [Google Scholar]
- Brou C, Kuhn A, Staub A, Chaudhary S, Grummt I, Davidson I, Tora L (1993) Sequence-specific transactivators counteract topoisomerase II-mediated inhibition of in vitro transcription by RNA polymerases I and II. Nucleic Acids Res 21: 4011–4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Belmont AS, Huang S (2004) Upstream binding factor association induces large-scale chromatin decondensation. Proc Natl Acad Sci USA 101: 15106–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L, Tanese N, Tjian R (1992) The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell 68: 965–976 [DOI] [PubMed] [Google Scholar]
- Comai L, Zomerdijk JC, Beckmann H, Zhou S, Admon A, Tjian R (1994) Reconstitution of transcription factor SL1: exclusive binding of TBP by SL1 or TFIID subunits. Science 266: 1966–1972 [DOI] [PubMed] [Google Scholar]
- Friedrich JK, Panov KI, Cabart P, Russell J, Zomerdijk JC (2005) TBP–TAF complex SL1 directs RNA polymerase I pre-initiation complex formation and stabilizes upstream binding factor at the rDNA promoter. J Biol Chem 280: 29551–29558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JA, Tjian R (1994) TBP–TAF complexes: selectivity factors for eukaryotic transcription. Curr Opin Cell Biol 6: 403–409 [DOI] [PubMed] [Google Scholar]
- Grummt I (1999) Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog Nucleic Acid Res Mol Biol 62: 109–154 [DOI] [PubMed] [Google Scholar]
- Grummt I (2003) Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev 17: 1691–1702 [DOI] [PubMed] [Google Scholar]
- Hahn S (1998) The role of TAFs in RNA polymerase II transcription. Cell 95: 579–582 [DOI] [PubMed] [Google Scholar]
- Hannan KM, Hannan RD, Rothblum LI (1998) Transcription by RNA polymerase I. Front Biosci 3: d376–d398 [DOI] [PubMed] [Google Scholar]
- Heix J, Grummt I (1995) Species specificity of transcription by RNA polymerase I. Curr Opin Genet Dev 5: 652–656 [DOI] [PubMed] [Google Scholar]
- Heix J, Zomerdijk J, Ravanpay A, Tjian R, Grummt I (1997) Cloning of murine RNA polymerase I-specific TAF factors: conserved interactions between the subunits of the species-specific transcription initiation factor TIF-IB/SL1. Proc Natl Acad Sci USA 94: 1733–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N (1993) TBP, a universal eukaryotic transcription factor? Genes Dev 7: 1291–1308 [DOI] [PubMed] [Google Scholar]
- Hisatake K, Nishimura T, Maeda Y, Hanada K, Song CZ, Muramatsu M (1991) Cloning and structural analysis of cDNA and the gene for mouse transcription factor UBF. Nucleic Acids Res 19: 4631–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James MJ, Zomerdijk JC (2004) Phosphatidylinositol 3-kinase and mTOR signaling pathways regulate RNA polymerase I transcription in response to IGF-1 and nutrients. J Biol Chem 279: 8911–8918 [DOI] [PubMed] [Google Scholar]
- Jantzen HM, Admon A, Bell SP, Tjian R (1990) Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature 344: 830–836 [DOI] [PubMed] [Google Scholar]
- Jantzen HM, Chow AM, King DS, Tjian R (1992) Multiple domains of the RNA polymerase I activator hUBF interact with the TATA-binding protein complex hSL1 to mediate transcription. Genes Dev 6: 1950–1963 [DOI] [PubMed] [Google Scholar]
- Kiss AM, Jady BE, Bertrand E, Kiss T (2004) Human box H/ACA pseudouridylation guide RNA machinery. Mol Cell Biol 24: 5797–5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Grummt I (1992) Dual role of the nucleolar transcription factor UBF: trans-activator and antirepressor. Proc Natl Acad Sci USA 89: 7340–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H, Green MR (1994) The RNA polymerase I transcription factor, upstream binding factor, interacts directly with the TATA box-binding protein. J Biol Chem 269: 30140–30146 [PubMed] [Google Scholar]
- Mais C, Wright JE, Prieto JL, Raggett SL, McStay B (2005) UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev 19: 50–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G, Panov KI, Friedrich JK, Trinkle-Mulcahy L, Lamond AI, Zomerdijk JC (2001) hRRN3 is essential in the SL1-mediated recruitment of RNA polymerase I to rRNA gene promoters. EMBO J 20: 1373–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T (2004) At the crossroads of growth control; making ribosomal RNA. Curr Opin Genet Dev 14: 210–217 [DOI] [PubMed] [Google Scholar]
- Moss T, Stefanovsky VY (2002) At the center of eukaryotic life. Cell 109: 545–548 [DOI] [PubMed] [Google Scholar]
- Nomura M (2001) Ribosomal RNA genes, RNA polymerases, nucleolar structures, and synthesis of rRNA in the yeast Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol 66: 555–565 [DOI] [PubMed] [Google Scholar]
- O'Mahony DJ, Rothblum LI (1991) Identification of two forms of the RNA polymerase I transcription factor UBF. Proc Natl Acad Sci USA 88: 3180–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan AC, Sullivan GJ, McStay B (2002) UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol Cell Biol 22: 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panov KI, Friedrich JK, Russell J, Zomerdijk JC (2006) UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J 25: 3310–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panov KI, Friedrich JK, Zomerdijk JC (2001) A step subsequent to preinitiation complex assembly at the ribosomal RNA gene promoter is rate limiting for human RNA polymerase I-dependent transcription. Mol Cell Biol 21: 2641–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panova TB, Panov KI, Russell J, Zomerdijk JC (2006) Casein kinase 2 associates with initiation-competent RNA polymerase I and has multiple roles in ribosomal DNA transcription. Mol Cell Biol 26: 5957–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier G, Stefanovsky VY, Faubladier M, Hirschler-Laszkiewicz I, Savard J, Rothblum LI, Cote J, Moss T (2000) Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol Cell 6: 1059–1066 [DOI] [PubMed] [Google Scholar]
- Reeder RH (1999) Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog Nucleic Acid Res Mol Biol 62: 293–327 [DOI] [PubMed] [Google Scholar]
- Rudloff U, Eberhard D, Tora L, Stunnenberg H, Grummt I (1994) TBP-associated factors interact with DNA and govern species specificity of RNA polymerase I transcription. EMBO J 13: 2611–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J, Zomerdijk JC (2005) RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci 30: 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp A, Grummt I (1991) Transcription complex formation at the mouse rDNA promoter involves the stepwise association of four transcription factors and RNA polymerase I. J Biol Chem 266: 24588–24595 [PubMed] [Google Scholar]
- Smith SD, O'Mahony DJ, Kinsella BT, Rothblum LI (1993) Transcription from the rat 45S ribosomal DNA promoter does not require the factor UBF. Gene Expression 3: 229–236 [PMC free article] [PubMed] [Google Scholar]
- Smith SD, Oriahi E, Lowe D, Yang-Yen HF, O'Mahony D, Rose K, Chen K, Rothblum LI (1990) Characterization of factors that direct transcription of rat ribosomal DNA. Mol Cell Biol 10: 3105–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky V, Langlois F, Gagnon-Kugler T, Rothblum LI, Moss T (2006) Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol Cell 21: 629–639 [DOI] [PubMed] [Google Scholar]
- Wang L, Bhattacharyya N, Chelsea DM, Escobar PF, Banerjee S (2004) A novel nuclear protein, MGC5306 interacts with DNA polymerase beta and has a potential role in cellular phenotype. Cancer Res 64: 7673–7677 [DOI] [PubMed] [Google Scholar]
- Yuan X, Zhao J, Zentgraf H, Hoffmann-Rohrer U, Grummt I (2002) Multiple interactions between RNA polymerase I, TIF-IA and TAFI subunits regulate preinitiation complex assembly at the ribosomal gene promoter. EMBO Rep 3: 1082–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomerdijk JCBM, Beckmann H, Comai L, Tjian R (1994) Assembly of transcriptionally active RNA polymerase I initiation factor SL1 from recombinant subunits. Science 266: 2015–2018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Information


