Abstract
Argonautes (AGOs) are conserved proteins that contain an RNA-binding PAZ domain and an RNase H-like PIWI domain. In Arabidopsis, except for AGO1, AGO4 and AGO7, the roles of seven other AGOs in gene silencing are not known. We found that a mutation in AGO6 partially suppresses transcriptional gene silencing in the DNA demethylase mutant ros1-1. In ago6-1ros1-1 plants, RD29A promoter short interfering RNAs (siRNAs) are less abundant, and cytosine methylation at both transgenic and endogenous RD29A promoters is reduced, compared to that in ros1-1. Interestingly, the ago4-1 mutation has a stronger suppression of the transcriptional silencing phenotype of ros1-1 mutant. Analysis of cytosine methylation at the endogenous MEA-ISR, AtREP2 and SIMPLEHAT2 loci revealed that the CpNpG and asymmetric methylation levels are lower in either of the ago6-1 and ago4-1 single mutants than those in the wild type, and the levels are the lowest in the ago6-1ago4-1 double mutant. These results suggest that AGO6 is important for the accumulation of specific heterochromatin-related siRNAs, and for DNA methylation and transcriptional gene silencing, this function is partly redundant with AGO4.
Keywords: Argonaute, RNA-directed DNA methylation, silencing, small RNA
Introduction
Gene silencing occurs at the transcriptional and post-transcriptional levels (Baulcombe, 2004; Bender, 2004; Mello and Conte, 2004; Chan et al, 2005; Matzke and Birchler, 2005; Morris, 2005; Wassenegger, 2005). Post-transcriptional gene silencing (PTGS) involves microRNAs (miRNAs) and certain classes of short interfering RNAs (siRNAs) (Carrington and Ambros, 2003; Bartel, 2004; Baulcombe, 2004; He and Hannon, 2004; Tang, 2005). miRNAs are about ∼21–22 nt in size and are processed by Dicer family of endonuclease III enzymes from longer precursor RNAs that can form stem-loop structures (Carrington and Ambros, 2003; Bartel, 2004). siRNAs are generated from long double-stranded RNAs (dsRNAs) by enzymes of the Dicer family (Baulcombe, 2004; Matzke and Birchler, 2005; Bouche et al, 2006; Moissiard and Voinnet, 2006). There are two major size classes of siRNAs, that is, ∼21- and ∼24-nt siRNAs (Brodersen and Voinnet, 2006). miRNAs and siRNAs are incorporated into RNA-induced silencing complex (RISC) and guide the complex to complementary mRNAs, causing translational inhibition or transcript cleavage (He and Hannon, 2004; Meister and Tuschl, 2004; Tang, 2005). siRNAs of the 24-nt size classes can also guide DNA methylation and histone methylation to cause transcriptional gene silencing (TGS) (Baulcombe, 2004; Bender, 2004; Mello and Conte, 2004; Chan et al, 2005; Matzke and Birchler, 2005; Morris, 2005).
The PAZ and PIWI domain-containing Argonaute (AGO) proteins are the catalytic engine of RISC in PTGS (He and Hannon, 2004; Meister and Tuschl, 2004). In TGS, AGO is also a key component of RNA-induced transcriptional silencing complex (Verdel and Moazed, 2005) or of a hypothetical complex responsible for RNA-directed DNA methylation (RdDM) (Chan et al, 2005; Matzke and Birchler, 2005). In Schizosaccharomyces pombe, a single AGO protein mediates both transcriptional and post-transcriptional silencing (Sigova et al, 2004). In Caenorhabditis elegans, mutants defective in a member of the AGO gene family, RDE-1, are strongly resistant to RNA interference (RNAi) (Tabara et al, 1999). Another PAZ/PIWI protein, PPW-1, is required for efficient germline RNAi (Tijsterman et al, 2002). Systematic analysis of AGO mutants in C. elegans showed that distinct AGOs act sequentially during RNAi (Yigit et al, 2006). In Drosophila, AGO2 is directly involved in RISC formation as ‘Slicer' of the passenger strand of the siRNA duplex (Rand et al, 2005). AGO1, another member of the Drosophila AGO family, immunopurified from Schneider2 cells associates with miRNAs and cleaves target RNAs that are fully complementary to the miRNAs (Miyoshi et al, 2005). PIWI has been shown to be necessary for PTGS and some aspects of TGS (Pal-Bhadra et al, 2002). In humans, AGO2 (hAgo2) was shown to be responsible for target RNA cleavage (‘Slicer') activity in RNAi (Meister et al, 2004). In mouse, AGO2 contributes ‘Slicer' activity to RISC, providing the catalytic engine for RNAi (Liu et al, 2004).
In Arabidopsis, there are 10 predicted AGO proteins (Morel et al, 2002). AGO1 was first found as a regulator of Arabidopsis leaf development (Bohmert et al, 1998). AGO1 is also involved in PTGS and viral resistance (Morel et al, 2002; Zhang et al, 2006). It functions as an RNA slicer that selectively recruits miRNAs and some siRNAs (Vaucheret et al, 2004; Ronemus et al, 2006). AGO4 controls locus-specific siRNA accumulation and DNA methylation (Zilberman et al, 2003, 2004). Maintenance of the heterochromatic state involves locus-specific Pol IVa function followed by siRNA production and assembly of AGO4- and NRPD1b-containing silencing complex within nucleolar processing centers (Pontes et al, 2006). AGO4 interacts with the C-terminal domain of NRPD1b, and its stability depends on upstream factors that synthesize siRNAs (Li et al, 2006; Pontes et al, 2006). AGO4 can function at target loci through two distinct and separable mechanisms. First, AGO4 can recruit components that signal DNA methylation in a manner independent of its catalytic activity. Second, through the catalytic activity of AGO4, secondary siRNAs are generated to reinforce silencing (Qi et al, 2006). ZIPPY (AGO7) functions in the regulation of developmental timing and is needed for the production and/or stability of some trans-acting siRNAs (Hunter et al, 2003, 2006; Adenot et al, 2006; Fahlgren et al, 2006; Garcia et al, 2006). PINHEAD/ZWILLE (AGO10) plays a critical role in maintaining undifferentiated stem cells in the shoot apical meristem, but was found not to participate in PTGS (Moussian et al, 1998; Lynn et al, 1999; Morel et al, 2002). The functional roles of other AGO proteins in Arabidopsis remain to be determined.
In this study, we characterized a second site mutation that suppresses TGS in the Arabidopsis ros1-1 mutant. ROS1 is a 5-methylcytosine DNA glycosylase/lyase required for preventing DNA hypermethylation. In ros1 mutants, TGS occurs at the RD29A-LUC (firefly luciferase reporter gene driven by the stress-responsive RD29A promoter) transgene and the linked kanamycin -resistance gene 35S-NPTII (neomycin phosphotransferase II driven by the CaMV 35S promoter), and at the endogenous RD29A gene. Both the transgene and endogenous RD29A promoters are hypermethylated, which may be triggered by siRNAs produced from the transgene RD29A promoter (Gong et al, 2002). The suppressor mutation can partially release TGS at both transgene and endogenous RD29A promoters, but not at the 35S-NPTII transgene. In ago6-1ros1-1 double mutant, the cytosine methylation levels at both transgenic and endogenous RD29A promoters are reduced, and the amount of siRNAs generated from RD29A promoter region is much less than in ros1-1 plants. The suppressor mutation also reduces the levels of siRNAs and DNA methylation at some endogenous loci. The suppressor mutation was found to be in AGO6, a member of the AGO family. In addition, we found that the ago4-1 mutation can have a more complete suppression of ros1, compared to ago6-1. DNA methylation analysis of several endogenous loci in the ago4ago6 double mutant showed that the effect of the double mutant is stronger than either of the single mutants. Our results suggest that AGO6 has a partially redundant function with AGO4 in siRNA accumulation and in controlling DNA methylation and TGS at specific loci.
Results
Partial suppression of transgene and endogenous RD29A promoter transcriptional silencing in ros1-1 by the ago6-1 mutation
We screened for suppressors of ros1-1 from a T-DNA-mutagenized population (Kapoor et al, 2005) based on the enhanced bioluminescence compared with that in ros1-1 mutant. One of the suppressor mutants with increased bioluminescence (Figure 1A, C and D) was designated as ago6-1. The ago6-1 mutation, however, did not release the TGS of 35S-NPTII (Figure 1B), because the ros1-1ago6-1 plants are kanamycin -sensitive. This is in contrast to the rpa2 mutant (Kapoor et al, 2005), which can release the silencing of 35S-NPTII but not the linked RD29A-LUC in the ros1-1 mutant background. The results from rpa2 mutant studies suggested that the TGS of 35S-NPTII in ros1-1 mutant is not caused by siRNAs and is also not dependent on DNA methylation (Kapoor et al, 2005).
Figure 1.
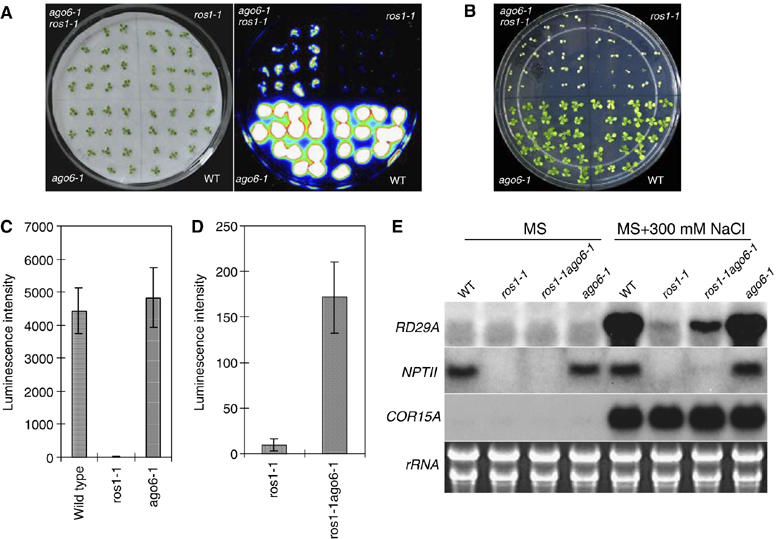
Partial suppression of RD29A-LUC and endogenous RD29A silencing but not of 35S-NPTII silencing in ros1 by ago6. (A) Luminescence image of WT, ros1-1, ros1-1ago6-1 and ago6-1. Luminescence imaging was taken with 12-day-old seedlings treated with 300 mM NaCl for 5 h. (B) Phenotype of WT, ros1-1, ros1-1ago6-1 and ago6-1 plants plated on MS medium supplemented with kanamycin (50 μg/ml). (C, D) Quantification of the luminescence intensities of WT, ros1-1, ros1-1ago6-1 and ago6-1 plants treated with 300 mM NaCl for 5 h. (E) Northern blot analysis of the transcript levels of NPT II, endogenous RD29A and the stress-responsive control gene COR15A in WT, ros1-1, ros1-1ago6-1 and ago6-1. WT, wild type of the C24 ecotype carrying a LUCIFERASE gene driven by the RD29A promoter (C24 RD29A-LUC).
Northern blot analysis revealed that in ros1-1ago6-1 double-mutant plants, the endogenous RD29A transcript accumulated to a level higher than in ros1-1 but the level was not as high as in the wild-type (WT) plants, indicating that the suppression of the endogenous RD29A gene silencing in ros1-1 is partial, as is the suppression of the RD29A-LUC transgene. However, the ros1-1ago6-1 mutant did not accumulate any NPTII transcripts, which is consistent with the kanamycin-sensitive phenotype of these plants (Figure 1E), and suggests that there is no suppression of the 35S-NPTII transgene gene silencing.
DNA methylation level at the RD29A promoter in ros1-1 is reduced by the ago6 -1 mutation
In ros1-1 mutant plants, DNA hypermethylation in the promoter region of both the RD29A-LUC transgene and the endogenous RD29A gene causes TGS at the two loci (Gong et al, 2002). We hypothesized that, in ros1-1ago6-1 plants, the ago6-1 mutation suppresses the TGS in ros1-1 probably by reducing the levels of DNA methylation at the promoter regions. Bisulfite sequencing was carried out to determine the DNA methylation levels at both the transgene and endogenous RD29A promoter regions. As shown in Figure 2A and B, and in Supplementary Figure 1A and B, DNA methylation levels at CpG, CpNpG and asymmetric sites of the transgene RD29A promoter were decreased in ros1-1ago6-1 compared with those in ros1-1 plants. At the endogenous RD29A promoter, a similar pattern of reduced methylation was found at CpG, CpNpG and asymmetric sites in the ros1-1ago6-1 double mutant, compared to ros1-1. These results support our notion that decreased DNA methylation levels at the transgene and endogenous RD29A promoters resulted in the suppression of ros1-1 by ago6-1.
Figure 2.
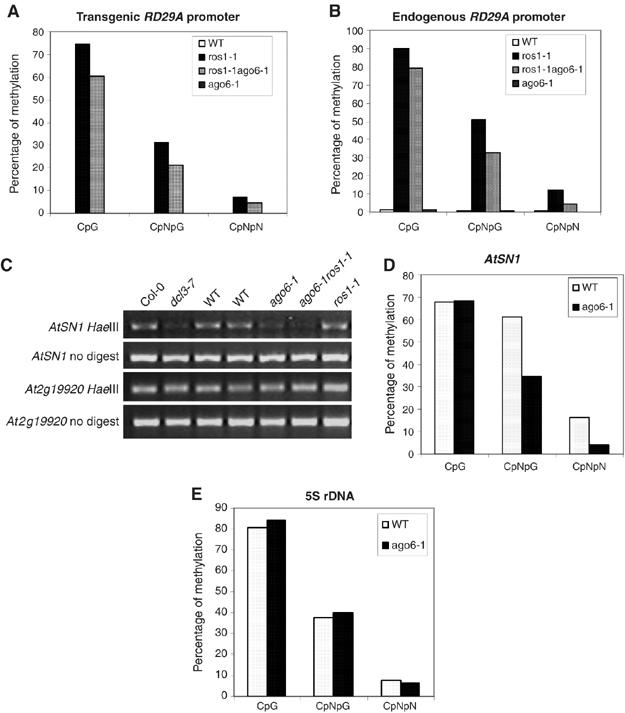
The effect of AGO6 on DNA methylation levels. (A) DNA methylation levels (percent of methylated DNA) of transgenic RD29A promoter, and (B) DNA methylation levels of endogenous RD29A promoter in WT, ros1-1, ros1-1ago6-1 and ago6-1. (C) ago6-1 causes decreased AtSN1 cytosine methylation. PCR was used to amplify a portion of the AtSN1 retro-element. Undigested DNA and a gene lacking HaeIII sites served as PCR controls. (D, E) DNA methylation levels at AtSN1 and 5S rDNA in WT and ago6-1.
The ago6-1 mutation reduces non-CpG methylation at specific endogenous loci
To determine whether the ago6-1 mutation affects the DNA methylation of endogenous sequences not associated with the transgene, we analyzed DNA methylation at the AtSN1 locus. First, a PCR-based method was used to determine whether ago6-1 mutation affects DNA methylation level of the retrotransposon AtSN1. The abundance of PCR products amplified from ago6-1 and ros1-1ago6-1 plants was much lower than that from ros1-1 and WT plants (Figure 2C), suggesting that the DNA methylation levels at the AtSN1 locus in ago6-1 and ros1-1ago6-1 plants are lower than those in ros1-1 and WT plants. To confirm the results obtained by the PCR-based method, we used bisulfite sequencing. Although CpG methylation levels were unchanged, ago6-1 showed almost two-fold reduction in CpNpG and four-fold reduction in asymmetric methylation at the AtSN1 locus (Figure 2D and Supplementary Figure 2A). We also carried out bisulfite sequencing of the 5S rDNA repeat. The results showed that there was no substantial reduction in CpG, CpNpG or asymmetric methylation in the ago6-1 mutant (Figure 2E and Supplementary Figure 2B). These results suggest that ago6 affects CpNpG and asymmetric methylation at some specific endogenous loci.
The ago6-1 mutation reduces the accumulation of heterochromatin-related siRNAs from transgene and endogenous loci
In ros1-1 plants, DNA hypermethylation at the transgene and endogenous RD29A promoters may be triggered by siRNAs produced from the transgene promoter (Gong et al, 2002). It is possible that the reduced DNA methylation levels at the RD29A promoter regions in ros1-1ago6-1 double mutant are a result of decreased accumulation or activity of the promoter siRNAs. Therefore, Northern blot analysis was carried out to assess the siRNA levels. The 24-nt siRNAs produced from the RD29A promoter were less abundant in ago6-1 than that in the WT (Figure 3A), and similarly, the siRNA level was also less abundant in ros1-1ago6-1 than that in ros1-1 plants (Figure 3B). The siRNAs were absent in plants that do not contain the RD29A-LUC transgene, such as in Col-0, dcl3-7, rdr2-1 and ago6-2 plants (Figure 3A).
Figure 3.
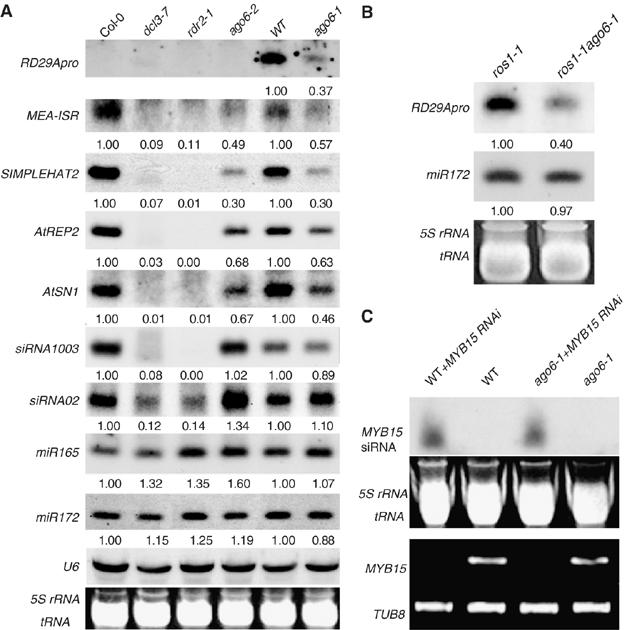
Effect of ago6 on the accumulation of small RNAs. (A) Effect of ago6 on the accumulation of siRNAs from the transgenic RD29A promoter, endogenous siRNAs and miRNAs. In addition to ago6-1, a SALK T-DNA insertion allele, ago6-2 and its WT (Col-0) were also analyzed. snoRNA U6 and ethidium bromide-stained tRNA and rRNA bands served as loading controls. Relative levels of the small RNAs were calculated and shown below the small RNA bands. (B) The transgene RD29A promoter siRNA was less abundant in ros1-1ago6-1 double compared with that in the ros1-1 single mutant. (C) The effect of ago6 on the accumulation of siRNAs from transgenic inverted repeat MYB15 dsRNA. MYB15 transcript levels were analyzed by RT–PCR.
The accumulation pattern of several endogenous siRNAs was also investigated (Figure 3A). AtSN1 and MEA-ISR siRNAs were detected in both ago6-1 and ago6-2, but the levels were much lower than those in their respective WT backgrounds (Col-0 for ago6-2, and WT (C24 containing RD29A-LUC) for ago6-1). AtREP2 and SIMPLEHAT2 siRNA levels were also decreased in both ago6-1 and ago6-2 mutants compared to those in their respective WT. The levels of siRNA1003 were not substantially different in ago6-1 or ago6-2, compared to their respective WT controls. The ago6 mutations also did not reduce the accumulation of siRNA02 (Figure 3A).
It has been shown that AGO1 is physically associated with miRNAs, but not with virus-derived siRNAs and 24-nt siRNAs involved in chromatin silencing (Baumberger and Baulcombe, 2005). We determined the levels of miRNAs to determine whether ago6 mutations have any effect on miRNA accumulation. As shown in Figure 3A, there was no substantial reduction in miR165 or mi172 levels in any of the mutants compared to their respective WT controls. The above results show that AGO6 is partially required for the accumulation of certain 24-nt chromatin-related siRNAs.
The ago6-1 mutation does not affect PTGS caused by inverted repeat transgenes
PTGS of endogenous genes mediated by introduction of dsRNA was demonstrated in various organisms (Mello and Conte, 2004). To determine whether ago6-1 affects siRNAs from hairpin constructs targeting mRNAs for cleavage, we crossed an inverted repeat construct of the Arabidopsis MYB15 gene under the control of CaMV 35S promoter (Agarwal et al, 2006) to the ago6-1 mutant background. Accumulation of siRNAs derived from the MYB15 dsRNA construct and the transcript level of MYB15 gene were detected by Northern blot and reverse transcription (RT)–PCR analyses, respectively. As shown in Figure 3C, the MYB15 siRNA level in ago6-1 plants was not substantially different from that in the WT. The result suggests that AGO6 is not required for the accumulation of siRNAs derived from PTGS inverted repeat constructs. Consistent with this, the expression of the target gene MYB15 was silenced in ago6-1 mutant, as in the WT, again indicating that AGO6 does not function in the RNAi pathway for PTGS.
Cloning and characterization of the AGO6 gene
To determine the T-DNA insertion site in the ros1-1ago6-1 plants, thermal asymmetric interlaced PCR (TAIL-PCR) was carried out and an insertion was found in the 14th exon of the gene At2G32940, which is annotated as AGO6. This mutant allele was thus designated as ago6-1 (Figure 4A). Another allele of ago6 was obtained from the SALK T-DNA collection (Salk_031553), and was designated as ago6-2 (Figure 4A). To determine whether the gene At2G32940 was responsible for the phenotype of TGS suppression of ros1-1, a cosegregation analysis was conducted. A cross was made between ros1-1ago6-1 double mutant and ros1-1 single mutant, and its F1 was then selfed to generate F2, from which 35 individual plants with enhanced bioluminescence were selected for PCR analysis to determine their genotype at the AGO6 locus. The result showed that all of the 35 plants had a homozygous ago6-1 insertion, suggesting that the ago6-1 gene is responsible for the phenotype of enhanced luminescence in the ros1-1ago6-1 plants.
Figure 4.
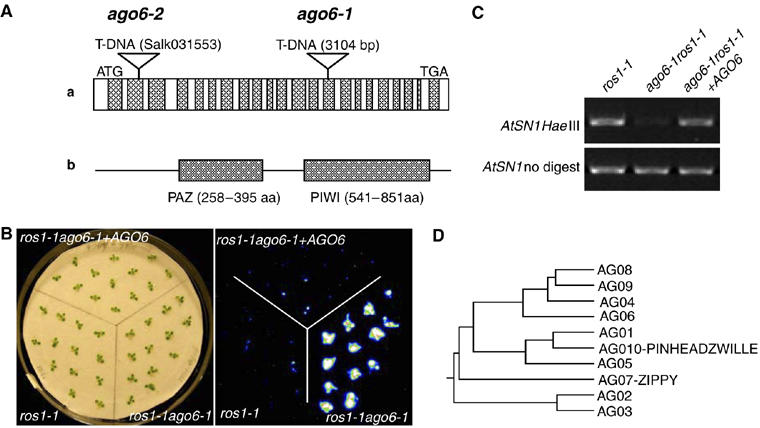
AGO6 gene cloning and diagram of structure. (A) AGO6 gene (At2G32940) structure. AGO6 gene has a total of 22 exons and encodes a putative AGO protein containing a PAZ domain and a PIWI domain. ago6-1 has a T-DNA insertion in the 14th exon, and ago6-2 (a T-DNA line from the SALK collection) (Salk 031553) has an insertion in the second exon. (B) Complementation of ago6-1 mutant. ros1-1ago6-1 plants transformed with WT AGO6 transgene were restored in the luminescence phenotype to that seen in ros1-1. (C) The molecular phenotype of AtSN1 methylation in ros1-1ago6-1 was restored after the ros1-1ago6-1 double mutant was transformed with the WT AGO6 gene. (D) Phylogenetic tree of the Arabidopsis AGO proteins using full-length amino-acid sequences. AGO4, 6, 8 and 9 clustered together.
To confirm further that At2G32940 is the correct gene, its genomic DNA including 1143 bp upstream of the initiation codon and 1533 bp downstream of the stop codon was cloned by PCR amplification into the binary vector pCAMBIA1305.1, which was then introduced into ros1-1ago6-1 plants by Agrobacterium-mediated transformation. T2 transgenic plants were subjected to phenotypic analysis. As shown in Figure 4B, a comparison of luminescence intensity of ros1-1 single mutant, ros1-1ago6-1 double mutant and ros1-1ago6-1 double mutant expressing the AGO6 transgene demonstrated that the AGO6 transgene construct restored the luminescence in ros1-1ago6-1 to the level seen in ros1-1. Thus, the mutant was complemented, which confirms that AGO6 is the correct gene (Figure 4B). We also used a PCR-based method to test the DNA methylation level at the AtSN1 locus in ros1-1, ros1-1ago6-1 and ros1-1ago6-1 carrying the AGO6 transgene. As shown in Figure 4C, in ros1-1ago6-1 plants that contain the AGO6 transgene, the DNA methylation level was restored to that in ros1-1 plants, further confirming that ago6 is responsible for the mutant phenotypes.
AGO6 encodes an 879-amino-acid protein that contains conserved PAZ and PIWI domains, characteristic of the AGO protein family. Arabidopsis contains 10 AGO proteins (Morel et al, 2002), and the available evidence suggests that each member may function differently. AGO1 and AGO4 were shown to participate in PTGS and TGS, respectively (Zilberman et al, 2003; Baumberger and Baulcombe, 2005). ZIPPY(AGO7) and PINHEAD/ZWILLE (AGO10) were found to have important roles in plant development (Moussian et al, 1998; Morel et al, 2002; Hunter et al, 2003). Phylogenetic analysis of full-length amino-acid sequences reveled that the 10 Arabidopsis AGO proteins fall into four clusters (Figure 4D). The AGO1, AGO5 and AGO10/PINHEAD/ZWILLE clade has high-sequence homologies with the human slicer AGO2, which was shown to target RNA for cleavage in the RNAi pathway (Meister et al, 2004). Indeed, recent studies showed that the Arabidopsis AGO1 is a bona fide Slicer (Baumberger and Baulcombe, 2005; Qi et al, 2005). AGO6 is in the same clade with AGO4, AGO8 and AGO9. AGO4 controls locus-specific siRNA accumulation and regulates DNA methylation (Zilberman et al, 2003, 2004). Our results, which that AGO6 also functions in siRNA accumulation and DNA methylation associated with TGS, is consistent with the phylogenetic clustering of AGO4 and AGO6. It is possible that the other two members of this cluster, AGO8 and AGO9 also function in chromatin siRNA accumulation and DNA methylation.
To determine the subcellular localization of AGO6 protein, a translational fusion between yellow fluorescence protein (YFP) and the N terminus of AGO6 was carried out. YFP-AGO6 protein was found to be localized mainly in the nucleus and weakly outside the nuclear compartment (Figure 5A). A construct of AGO6 genomic fragment containing an AGO6 C-terminal translational fusion to the MYC tag and driven by the endogenous AGO6 promoter was transformed into ros1-1ago6-1 double-mutant plants. This construct restored the RD29A-LUC silencing phenotype (data not shown). The T2 transgenic plants were subjected to immunostaining with the anti-MYC antibodies to confirm AGO6 protein localization. The result shows that the AGO6-MYC fusion protein was located in the nucleus (Figure 5Ba–c). In ros1-1ago6-1 mutant plants without the AGO6-MYC transgene, there was no immunostaining in the nucleus (Figure 5Bd–f). There did not appear to be a clear immunostaining in the nucleolus (Figure 5B). This predominant nuclear localization of AGO6 is in line with its function in chromatin siRNA accumulation, DNA methylation and transcriptional silencing.
Figure 5.
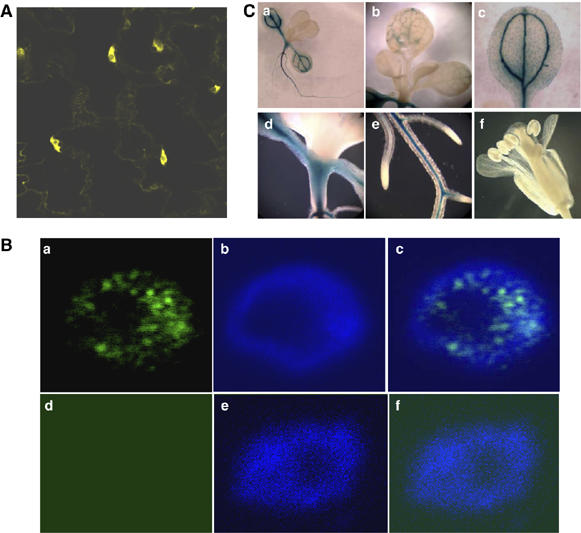
Subcellular localization of AGO6 protein and expression pattern of AGO6 promoter-driven GUS reporter gene. (A) Expression of YFP–AGO6 translational fusion under the control of CaMV 35S promoter in the epidermal cells of Arabidopsis. YFP–AGO6 protein was mainly localized in nuclei, and weakly in the cytoplasm. (B) Subcellular localization of AGO6 protein detected by immunostaining. (a) Localization of AGO6-MYC. (b) Nucleus with DAPI staining. (c) Overlap of (a) and (b). (d), (e) and (f) are negative controls showing immunostaining of a nucleus from ros1-1ago6-1 plant without the AGO6-MYC transgene. (C) AGO6 promoter–GUS expression pattern. AGO6 was strongly expressed in roots (a, e) and cotyledons (a–d), very weakly in young leaves (a, b) and not detectable in floral tissues (f).
An AGO6 promoter–GUS fusion was used to analyze AGO6 gene expression pattern. As shown in Figure 5C, AGO6 promoter–GUS expression was strong in roots, cotyledons and shoot meristematic region. The expression was weak in old and young leaves, and was not detectable in floral tissues.
Strong suppression of transcriptional silencing in ros1-1 by the ago4-1 mutation
As AGO4 also controls locus-specific siRNA accumulation and DNA methylation (Zilberman et al, 2003, 2004), we wondered whether ago4-1, like ago6-1, may suppress transgene and endogenous RD29A promoter transcriptional silencing in ros1-1. We crossed ago4-1 with ros1-1 and subsequently obtained ago4-1ros1-1 double-mutant plants. As shown in Figure 6A, ros1-1ago4-1 double-mutant seedlings emitted strong bioluminescence compared to that from the ros1-1 single mutant, indicating that ago4-1 strongly suppresses the TGS of RD29A-LUC in ros1-1 plants. We performed Northern analysis to determine if ago4-1 can also suppress the silencing of the endogenous RD29A gene in ros1-1. The result shows that the transcript level of endogenous RD29A in the ros1-1ago4-1 double mutant was nearly as high as that in WT plants (Figure 6B), indicating that ago4-1 also suppresses the TGS of endogenous RD29A in ros1-1.
Figure 6.
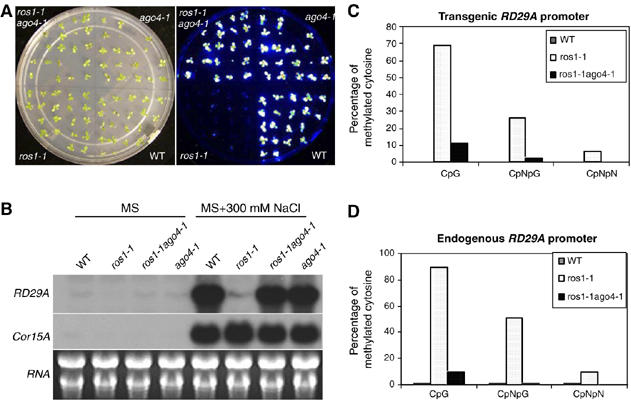
Strong suppression of RD29A-LUC and endogenous RD29A silencing in ros1-1 by ago4. (A) Luminescence images of WT, ros1-1, ros1-1ago4-1 and ago4-1 seedlings. Luminescence images were taken with 12-day-old seedlings treated with 300 mM NaCl for 5 h. (B) Northern blot analysis of the transcript levels of endogenous RD29A and the stress-responsive control gene COR15A in WT, ros1-1, ros1-1ago4-1 and ago4-1. (C) DNA methylation levels (percentage of methylated cytosine) of transgenic RD29A promoter, and (D) DNA methylation levels of endogenous RD29A promoter in WT, ros1-1 and ros1-1ago4-1.
To determine whether the suppression effect of ago4-1 is associated with changes in cytosine methylation, bisulfite sequencing was conducted at both transgene and endogenous RD29A promoters. We found that the CpG methylation level at transgene RD29A promoter was greatly reduced, and the methylation at CpNpG and asymmetric sites was almost lost in ros1-1ago4-1 compared to that in ros1-1 plants (Figure 6C and Supplementary Figure 3A). At the endogenous RD29A promoter, a similar pattern of reduced methylation at CpG, CpNpG and asymmetric sites was found in the ros1-1ago4-1 double mutant (Figure 6D and Supplementary Figure 3B). These results suggest that the suppression of ros1-1 by ago4-1 was caused by decreased DNA methylation levels at the transgene and endogenous RD29A promoters.
AGO6 functions redundantly with AGO4
We crossed ago6-1 and ago4-1 plants, and generated an ago6-1ago4-1 double mutant. DNA methylation levels at MEA-ISR, AtREP2, SIMPLEHAT2 and AtGP1 were analyzed in ago6-1 and ago4-1 single mutants, and in the ago6-1ago4-1 double mutant. At the MEA-ISR (Figure 7Aa and Supplementary Figure 4A), AtREP2 (Figure 7Ab and Supplementary Figure 4B) and SIMPLEHAT2 (Figure 7Ac and Supplementary Figure 5A–C) loci, there were substantial decreases in the mutants in DNA methylation at CpNpG and asymmetric sites but not at CpG sites. The results show that both AGO6 and AGO4 are important for CpNpG and asymmetric but not CpG cytosine methylation at the MEA-ISR, AtREP2 and SIMPLEHAT2 loci. These results are consistent with the observation that both ago6-1 (Figure 3A) and ago4-1 (Qi et al, 2006) affect MEA-ISR, AtREP2 and SIMPLEHAT2 siRNAs. Importantly, the results show that in the ago6-1ago4-1 double mutant, CpNpG and asymmetric cytosine methylation levels were even lower than those in ago6-1 or ago4-1 single mutant. In contrast, at the AtGP1 locus the methylation level at CpG, CpNpG and asymmetric sites did not show substantial differences among WT, ago6-1, ago4-1 and ago6-1ago4-1 (Figure 7Ad and Supplementary Figure 5D), suggesting that ago6-1 and ago4-1 do not affect the DNA methylation level at this locus.
Figure 7.
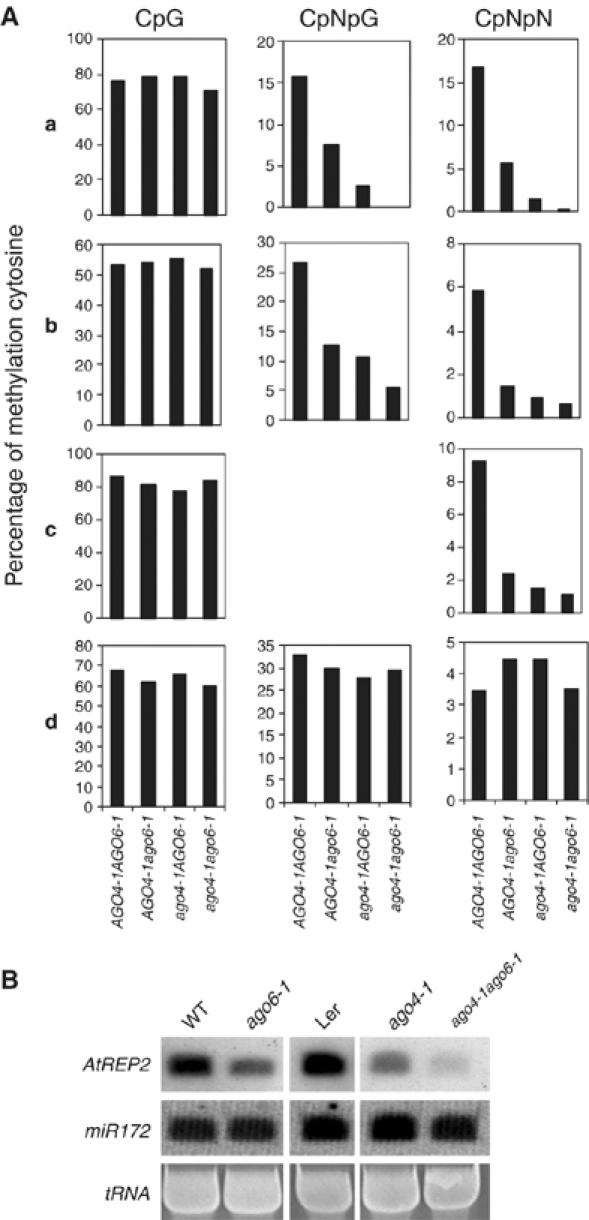
DNA methylation analysis at several endogenous loci in AGO4-1AGO6-1, AGO4-1ago6-1, ago4-1AGO6-1 and ago4-1ago6-1. (A) CpG (left), CpNpG (middle) and CpNpN (right) methylation at the MEA-ISR (a), AtREP2 (b), SIMPLEHAT2 (c) and AtGP1 (d) loci were analyzed by bisulfite sequencing. Methylation levels are shown by the percentage of methylated cytosine in all sequenced clones. Detailed bisulfite sequencing data are in Supplementary Figures. (B) Northern blot analysis of siRNAs at the AtREP2 locus in ago4-1, ago6-1 single mutants and ago4-1ago6-1 double mutant. miR172 was used as a control.
Northern blot analysis of AtREP2 siRNAs in ago4-1 and ago6-1 single mutants and ago4-1ago6-1 double mutant was carried out. As shown in Figure 7B, the AtREP2 siRNA levels in ago4-1, ago6-1 single mutants were lower than those in their respective WT controls. Furthermore, the ATREP2 siRNA level in ago4-1ago6-1 double mutant was lower than that in either ago4-1 or ago6-1 single mutant. The result is consistent with the effect of ago4-1ago6-1 on the DNA methylation at the AtREP2 locus. Taken together, these results suggest that the functions of AGO6 and AGO4 at some target loci are at least partly redundant.
Discussion
In this study, we identified the ago6-1 mutation as a suppressor of ros1-1. The ago6 mutation partially releases TGS of transgenic RD29A-LUC and endogenous RD29A gene, but not of the 35S-NPTII transgene caused by the ros1 mutation. Previously, we have reported that a mutation in the replication protein A2 suppresses the TGS of 35S-NPTII but not RD29A-LUC or endogenous RD29A, in a DNA methylation-independent manner (Kapoor et al, 2005). Our study here further supports that the mechanism of silencing at 35S-NPTII is different from that of silencing at the RD29A promoter.
Analyzing the DNA methylation pattern at both transgene and endogenous RD29A promoter regions revealed that in the ros1-1ago6-1 double mutant the levels of CpG, CpNpG and asymmetric methylation were all decreased compared to those in ros1-1 mutant. In addition, we found that ago6-1 mutation reduces DNA methylation at endogenous targets such as AtSN1, MEA-ISR, AtREP2 and SIMPLEHAT2, because the levels of CpNpG and asymmetric methylation were decreased in the ago6-1 mutant. Interestingly, CpG methylation levels at these endogenous loci were not lower in the ago6 mutant. Therefore, it seems that AGO6 is important for DNA methylation at all sequence context for some loci, but is only required for DNA methylation at non-CpG sites for some other loci.
siRNAs are believed to be the trigger for de novo DNA methylation in RdDM (Mello and Conte, 2004; Chan et al, 2005; Morris, 2005). Analysis of siRNA levels in ago6-1 mutant showed that siRNAs from the transgenic RD29A promoter was substantially reduced. The ago6-1 mutation also substantially decreased the levels of siRNAs from endogenous loci such as AtSN1, MEA-ISR, AtREP2 and SIMPLEHAT2, but did not reduce siRNA1003 and siRNA02 levels. These results demonstrate an important role of AGO6 in the accumulation of specific chromatin-related siRNAs. In addition, our results suggest that AGO6 does not have a substantial effect on miRNAs and PTGS siRNAs from inverted repeat transgenes. How AGO6 is involved in the accumulation of chromatin-related siRNAs remains an open question. It is possible that AGO6 functions in RdDM, and its role in siRNA accumulation is indirect and is perhaps through its effect on DNA methylation in a cycle of feed forward regulation involving transcription of methylated DNA to generate more siRNAs, similar to what has been proposed for AGO4 (Li et al, 2006; Pontes et al, 2006; Qi et al, 2006).
Arabidopsis has 10 members in the AGO family (Morel et al, 2002). AGO6 is in the same subfamily with AGO4, although within this subfamily AGO4 is more closely related in sequence to AGO8 and AGO9 than AGO6. AGO4 was first identified because of a partial suppression of epigenetic silencing at the Superman locus by ago4 mutations (Zilberman et al, 2003). The ago4 mutations decrease substantially non-CpG methylation but only affect slightly CpG methylation. The ago4 mutations were shown to block the accumulation of AtSN1, 5S rDNA, MEA-ISR, AtREP2 and SIMPLEHAT2 siRNAs, but had no effect on siRNA02 (Zilberman et al, 2003, 2004; Li et al, 2006; Pontes et al, 2006; Qi et al, 2006). It seems that AGO6 and AGO4 have very similar or overlapping functions. Compared to ago6, the effect of the ago4 mutations on siRNA accumulation and DNA methylation is stronger for some target loci such as the transgene and endogenous RD29A promoter regions and MEA-ISR. However, for some other target loci such as AtREP2 and SIMPLEHAT2, the ago6 and ago4 mutations have comparable effects. It is possible that ago6 may have a stronger effect than ago4 on some as yet unidentified target loci. Analysis of DNA methylation in ago4-1ago6-1 showed that non-CpG methylation at MEA-ISR, AtREP2 and SIMPLEHAT2 was lower in ago4-1ago6-1 compared to ago4-1 or ago6-1 single mutant, suggesting that the two AGO proteins may have partially redundant functions. This notion is also supported by the much reduced AtREP2 siRNAs in the ago4-1ago6-1 double mutant compared to ago4-1 or ago6-1 single mutant. Clearly, much further work is needed to determine the biochemical function of AGO6 in TGS and its relationship with other AGO family members. Notwithstanding, our results here provide strong evidence that AGO6 is important for the accumulation of siRNAs, DNA methylation and TGS at specific loci.
Materials and methods
Plant materials, mutant isolation and gene cloning
Unless specified otherwise, WT in this study refers to the C24 ecotype carrying a firefly luciferase reporter gene driven by the RD29A promoter (C24 RD29A-LUC) (Ishitani et al, 1997). A T-DNA insertion population in the ros1-1 background of Arabidopsis thaliana was generated as described (Kapoor et al, 2005). T2 seedlings were screened for ros1 suppressors on the basis of luminescence emission and kanamycin resistance. Luminescence imaging was carried out as described (Ishitani et al, 1997; Kapoor et al, 2005). T-DNA insertion sites were determined by using TAIL-PCR (Liu et al, 1995).
Total RNA and small RNA extractions and Northern blot analysis
Total RNA was extracted from 15-day-old seedlings by using 4 M guanidine thiocyanate (GT) extraction solution (4 M GT, 25 mM sodium citrate, 0.5% sarcosyl and 0.1 M mercaptoethanol). Briefly, 1–2 g of fresh material was ground in liquid N2 and powder was decanted into a 10 ml polypropylene tube that contained 3 ml cold 4 M GT solution. After shaking, 0.3 ml of 2 M NaAc (pH 5.0), 3 ml of acid phenol and 1 ml of chloroform were added in this order and mixed. After the mixture was centrifuged at 9000 r.p.m. for 12 min at 4°C, the supernatant was transferred to a new tube, 2 volumes of 100% ethanol was added and precipitated at −70°C for 1–1.5 h. The pellet was centrifuged at 6000 r.p.m. for 10 min, and suspended in 1 ml of 4 M LiCl. Then the tube was centrifuged at 13 000 r.p.m. for 12 min at 4°C, and the supernatant (containing small size RNAs) was transferred into a new 1.5 ml tube for later siRNA purification. The pellet (containing large size RNAs) was dissolved in 0.5 ml of DEPC-treated water and extracted with 0.5 volume of acid phenol and 0.5 volume of chloroform. The supernatant was transferred into a new tube after centrifuging at 13 000 r.p.m. for 10 min and then precipitated at −20°C for overnight by adding 0.1 volume of 3 M NaAc (pH 5.0) and 2 volumes of 100% ethanol. The pellet was centrifuged at 13 000 r.p.m. for 15 min and resuspended in RNase-free water.
For siRNA purification, the supernatant containing small size RNAs in 4 M LiCl was precipitated at −70°C for overnight by adding 0.1 volume of 3 M NaAc and equal volume of isopropanol. After being centrifuged at 13 000 r.p.m. for 20 min, the pellet was dried and dissolved in 500 μl of DEPC-treated water, and then extracted with 0.5 volume of acid phenol and 0.5 volume of chloroform. After centrifuging at 13 000 r.p.m. for 12 min, the supernatant was transferred to a new tube and precipitated at 4°C for 30 min after an equal volume of precipitate solution (1 M NaCl, 20% PEG8000) was added. After centrifuging at 13 000 r.p.m. for 10 min, the supernatant was transferred to a new tube, and precipitated at −70°C for overnight by adding 0.1 volume of NaAc (pH 5.0) and equal volume of isopropanol. The pellet was centrifuged at 13 000 r.p.m. for 20 min and resuspended in RNase-free water.
For analysis of mRNAs, 20 μg of total RNA was resolved on 1.2% denaturing agarose gel (MOPS-formaldehyde) and blotted onto nylon membrane. For small RNA analysis, 80 μg of small RNA was resolved on a 17% polyacrylamide-7 M urea gel and transferred electrophoretically to Hybond N_membranes (Amersham, Piscataway). PerfectHyb Plus Hybridization Buffer (Sigma, H7033-125ML) was used for mRNAs and siRNAs hybridizations according to the manufacture's instructions. Probes used for mRNA Northern blot were according to Kapoor et al (2005). The siRNA oligos for siRNA Northern blots for siRNA02, siRNA1003, AtSN1 (Cao et al, 2003; Zilberman et al, 2004), AtREP2, SIMPLEHAT2, MEA-ISR (Qi et al, 2006), mir165, miR172 and U6 are listed in Supplementary Table 1. Small RNA Northern blots were quantified after removing background: (small RNA(X)/U6(X))/(small RNA(Col-0 or WT)/U6(Col-0 or WT)), here X indicates different samples, such as Col-0, dcl3-7, rdr2-1, ago6-2, WT and ago6-1. Col-0 was control for dcl3-7, rdr2-1, ago6-2 and WT was control for ago6-1.
PCR-based AtSN1 cytosine methylation assay
PCR-based AtSN1 methylation assay was performed according to Onodera et al (2005). Briefly, after purified DNA was digested with HaeIII (or undigested for controls), DNA was used for each PCR. PCR conditions were 2 min at 94°C, followed by 35 cycles of 94°C for 30 s, 53°C for 30 s and 72°C for 30 s. Primer for AtSN1 and At2g19920 are listed in Supplementary Table 1.
DNA bisulfite sequencing analysis
DNA bisulfite sequencing was performed according to Frommer et al (1992) and Kapoor et al (2005) with slight modifications. Briefly, after digestion with EcoRV and purification by phenol:chloroform (1:1), 14 μl (500–800 ng) of genomic DNA was denatured at 94°C for 5 min, and then 0.84 μl of fresh 5 N NaOH was added, followed by incubation at 39°C for 20 min. A total of 120 μl of DNA sodium bisulfite treatment mixture, containing 102 μl of fresh 40.5% sodium bisulfite (pH 5.0, adjusted with 10 N NaOH) (Sigma, S-9000-500G), 3 μl of fresh 20 mM hydroquinone (Sigma, H9003-100G) and 14.84 μl of denatured DNA was prepared and subjected to PCR under the following conditions: first four cycles: 55°C for 3 h and 94°C for 5 min, followed by one cycle at 55°C for 3 h, and finally incubated at 4°C. Sodium bisulfite-treated DNA was purified with Wizard DNA Clean-up system (Promega, #A 7280) according to the manufacturer's instruction, and at the last step, DNA was dissolved with 200 μl of H2O at 65°C. The recollected DNA was incubated at 37°C for 20 min after 13 μl of 5 N NaOH was added, followed by precipitation at −80°C overnight by adding 210 μl of NH4Ac (pH 7.0) and 1200 μl of 100% ethanol. After centrifuging at 13 000 r.p.m. for 20 min, DNA was dissolving in 100 μl of water. PCR was performed with LA Taq polymerase (TaKaRa) with the following program: PCR mixture was first denatured at 94°C for 5 min, followed by 50 cycles: 94°C for 15 s, 54°C for 15 s and 72°C for 30 s, and last cycle at 72°C extended for 10 min. PCR products were cloned with pGEM-T Easy Vector System I (Promega, no. A1360) and transfected into DH10B competent cells. For each sample, at least 18 clones were sequenced.
Immunostaining
A construct containing AGO6 genomic DNA fused with MYC tag driven by endogenous AGO6 promoter was inserted into the pCAMBIA 1305.1 vector. The construct was transformed into ros1-1ago6-1 double-mutant plants, and the transgene was shown to restore the bioluminescence phenotype of ros1-1ago6-1 plants to that of ros1-1. The T2 plants were subjected to immunostaining analysis. Nuclei were prepared according to Bowler et al (2004), and stained with monoclonal anti-MYC antibodies (Clontech, 1:200 dilution). The staining patterns were visualized via Alexa Fluor 488- conjugated secondary antibodies (Molecular Probe; 1:2000 dilution) under a Leica fluorescence microscope equipped with proper filters.
Transgenic plants
AGO6 genomic DNA was amplified from BAC clone, T21L14, with Platium pfx DNA polymerase (Invitrogen, no. 11708-013) with forward primer, AGO6-gDNA-5F and reverse primer, AGO6-gDNA-3R, and PCR product was cloned into the SacI and HindIII sites of pCAMBIA 1305.1. AGO-MYC construct was constructed by inserting AGO6-MYC-tag into pCAMBIA 1305.1. AGO6 cDNA containing the entire open reading frame was amplified by RT–PCR with forward primer, AGO6-cDNA-5F, and reverse primer, AGO6-cDNA-3R. The amplified product was cloned into pDONR207 (Invitrogen) with BP clonase (Invitrogen) following the manufacturer's instructions. pEearlyGate104 was used for construct to express YFP-AGO6 fusion protein in Arabidopsis. For AGO6 promoter–GUS construct, AGO6 promoter was amplified from BAC clone, T21L14, with Platium pfx DNA polymerase (Invitrogen, no. 11708-013) with forward primer, pAGO6-5F, and reverse primer, pAGO6-3R and the PCR product was cloned into the PstI and EcoRI sites of pCAMBIA 1391Z. The constructs were introduced into Arabidopsis through floral-dip transformation with Agrobacterium GV301. Transgenic lines were selected based on hygromycine resistance for AGO6 complementation, and Basta resistance for YFP-AGO6 and AGO6 promoter–GUS constructs. For GUS staining, transgenic plants were incubated overnight at 37°C in GUS staining buffer (3 mM x-Gluc, 0.1 M sodium-phosphate buffer, pH 7, 0.1% Triton X-100 and, 8 mM β-mercaptoethanol), and then chlorophyll was removed by incubating the tissue in 80% EtOH at 80°C.
Reverse transcription–PCR
A total of 34 μl of the mixture including 5 μg of total RNA, 2 μl of 100 mM poly(dT)18 and 27 μl DEPC-treated water was denatured at 65°C for 10 min, and then annealed at room temperature for 10 min. For RT, the annealed mixture was mixed with the following components: 5 μl 10 × RT buffer, 5 μl 10 mM dNTP, 4 μl 0.1 M DTT, 1 μl reverse transcriptase (Promega), 1 μl RNase inhibitor (TaKaRa) and then incubated at 37°C for 1 h, followed by incubation at 70°C for 10 min. TUB8 was used as loading control.
Supplementary Material
Supplemental Figure Legends
Supplementary Figures
Supplementary Table 1
Acknowledgments
This work was supported by a National Institutes of Health grant R01GM070795 to J-K Zhu.
References
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H (2006) DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol 16: 927–932 [DOI] [PubMed] [Google Scholar]
- Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK (2006) A R2R3-type myb transcription factor is involved in the cold-regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281: 37636–37645 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Baulcombe D (2004) RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC (2005) Arabidopsis Argonaute1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J (2004) Chromatin-based silencing mechanisms. Curr Opin Plant Biol 7: 521–526 [DOI] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C (1998) AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J 17: 170–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche N, Lauressergues D, Gasciolli V, Vaucheret H (2006) An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J 25: 3347–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J (2004) Chromatin techniques for plant cells. Plant J 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Brodersen P, Voinnet O (2006) The diversity of RNA silencing pathways in plants. Trends Genet 22: 268–280 [DOI] [PubMed] [Google Scholar]
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE (2003) Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 13: 2212–2217 [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301: 336–338 [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6: 351–360 [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC (2006) Regulation of auxin response factor3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol 16: 939–944 [DOI] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA 89: 1827–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Collier SA, Byrne ME, Martienssen RA (2006) Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol 16: 933–938 [DOI] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK (2002) ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111: 803–814 [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531 [DOI] [PubMed] [Google Scholar]
- Hunter C, Sun H, Poethig RS (2003) The Arabidopsis heterochronic gene ZIPPY is an Argonaute family member. Curr Biol 13: 1734–1739 [DOI] [PubMed] [Google Scholar]
- Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutierrez-Nava M, Poethig SR (2006) Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133: 2973–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu JK (1997) Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9: 1935–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Agarwal M, Andreucci A, Zheng X, Gong Z, Hasegawa PM, Bressan RA, Zhu JK (2005) Mutations in a conserved replication protein suppress transcriptional gene silencing in a DNA-methylation-independent manner in Arabidopsis. Curr Biol 15: 1912–1918 [DOI] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE (2006) An Argonaute4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126: 93–106 [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK (1999) The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126: 469–481 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA (2005) RNAi-mediated pathways in the nucleus. Nat Rev Genet 6: 24–35 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T (2004) Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T (2004) Mechanisms of gene silencing by double-stranded RNA. Nature 431: 343–349 [DOI] [PubMed] [Google Scholar]
- Mello CC, Conte D Jr (2004) Revealing the world of RNA interference. Nature 431: 338–342 [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC (2005) Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev 19: 2837–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moissiard G, Voinnet O (2006) RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc Natl Acad Sci USA 103: 19593–19598 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Morel JB, Godon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H (2002) Fertile hypomorphic Argonaute (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV (2005) siRNA-mediated transcriptional gene silencing: the potential mechanism and a possible role in the histone code. Cell Mol Life Sci 62: 3057–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian B, Schoof H, Haecker A, Jurgens G, Laux T (1998) Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J 17: 1799–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS (2005) Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622 [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA (2002) RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell 9: 315–327 [DOI] [PubMed] [Google Scholar]
- Pontes O, Li CF, Nunes PC, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS (2006) The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 126: 79–92 [DOI] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ (2005) Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19: 421–428 [DOI] [PubMed] [Google Scholar]
- Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ (2006) Distinct catalytic and non-catalytic roles of Argonaute4 in RNA-directed DNA methylation. Nature 443: 1008–1012 [DOI] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X (2005) Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123: 621–629 [DOI] [PubMed] [Google Scholar]
- Ronemus M, Vaughn MW, Martienssen RA (2006) MicroRNA-Targeted and small interfering RNA-mediated mRNA degradation is regulated by Argonaute, Dicer, and RNA-dependent RNA polymerase in Arabidopsis. Plant Cell 18: 1559–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigova A, Rhind N, Zamore PD (2004) A single Argonaute protein mediates both transcriptional and posttranscriptional silencing in Schizosaccharomyces pombe. Genes Dev 18: 2359–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC (1999) The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99: 123–132 [DOI] [PubMed] [Google Scholar]
- Tang G (2005) siRNA and miRNA: an insight into RISCs. Trends Biochem Sci 30: 106–114 [DOI] [PubMed] [Google Scholar]
- Tijsterman M, Okihara KL, Thijssen K, Plasterk RH (2002) PPW-1, a PAZ/PIWI protein required for efficient germline RNAi, is defective in a natural isolate of C. elegans. Curr Biol 12: 1535–1540 [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crete P, Bartel DP (2004) The action of Argonaute1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Moazed D (2005) RNAi-directed assembly of heterochromatin in fission yeast. FEBS Lett 579: 5872–5878 [DOI] [PubMed] [Google Scholar]
- Wassenegger M (2005) The role of the RNAi machinery in heterochromatin formation. Cell 122: 13–16 [DOI] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC (2006) Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127: 747–757 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH (2006) Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev 20: 3255–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE (2003) Argonaute4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE (2004) Role of Arabidopsis Argonaute4 in RNA-directed DNA methylation triggered by inverted repeats. Curr Biol 14: 1214–1220 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure Legends
Supplementary Figures
Supplementary Table 1


