Abstract
The mouse homeobox gene Otx2 plays essential roles at each step and in every tissue during head development. We have previously identified a series of enhancers that are responsible for driving the Otx2 expression in these contexts. Among them the AN enhancer, existing 92 kb 5′ upstream, directs Otx2 expression in anterior neuroectoderm (AN) at the headfold stage. Analysis of the enhancer mutant Otx2ΔAN/− indicated that Otx2 expression under the control of this enhancer is essential to the development of AN. This study demonstrates that the AN enhancer is promoter-dependent and regulated by acetylated YY1. YY1 binds to both the AN enhancer and promoter region. YY1 is acetylated in the anterior head, and only acetylated YY1 can bind to the sequence in the enhancer. Moreover, YY1 binding to both of these two sites is essential to Otx2 expression in AN. These YY1 binding sites are highly conserved in AN enhancers in tetrapods, coelacanth and skate, suggesting that establishment of the YY1 regulation coincides with that of OTX2 function in AN development in an ancestral gnathostome.
Keywords: acetylation, anterior neuroectoderm, enhancer, Otx2 , YY1
Introduction
Otx2 is a mouse paired-type homeobox gene homologous to the Drosophila head gap gene otd (Simeone et al, 1992). Its expression is first found in the inner cell mass at the blastocyst stage. At the egg cylinder stage, Otx2 is detected in the epiblast and visceral endoderm. At the headfold stage, its expression characterizes the anterior mesendoderm and anterior neuroectoderm (AN). Subsequently, Otx2 expression is lost in the most rostral portion of the AN, and at the pharyngeal stage it is observed in caudal forebrain and midbrain with a sharp caudal boundary at the isthmus, but is not found in rostral forebrain including telencephalon and hypothalamus (Kurokawa et al, 2004a). Otx2 is also expressed in neural crest cells at the migratory phase (Kimura et al, 1997), and at later stages is specifically expressed in several sites in the nervous system, including cerebellum and sensory organs.
We have previously identified cis-regulatory elements that drive Otx2 expression in various contexts (Kimura et al, 1997, 2000; Kurokawa et al, 2004a, 2004b). Enhancers that regulate Otx2 expression in visceral endoderm, anterior mesendoderm and cephalic mesenchyme occupy a 1.8 kb region 5′ proximal to the translation start site (Kimura et al, 1997, 2000). In contrast, the enhancers that direct the expression in inner cell mass/epiblast (EP) and AN are encoded approximately −92 kb 5′ upstream (Kurokawa et al, 2004b). The EP enhancer requires a 2.3 kb region; the AN enhancer is an essential component of the EP enhancer and has no activity in epiblast by itself. The AN activity covers the entire anterior neuroectoderm at the presomite and early somite stages. In the enhancer mutant, Otx2ΔAN/−, the anterior neuroectoderm is initially induced at E7.5, but subsequently undergoes caudalization into Gbx2-positive metencephalon; no Emx2-positive telencephalon is formed (Kurokawa et al, 2004b).
The AN enhancer, however, loses its activity by E8.5, and the subsequent Otx2 expression in anterior neuroectoderm is regulated by the FM enhancer, located 75 kb 5′ upstream, and the FM2 enhancer at 115 kb 3′ downstream. Otx2 expression under the control of the FM and FM2 enhancers overlaps with Otx1 expression; it is not found in rostral forebrain, which corresponds to the future telencephalon and hypothalamus, and the caudal boundary of its activity is sharply delineated at the isthmus. The compound mutant of the Otx1 gene and FM and FM2 enhancers, Otx1−/−Otx2ΔFMΔFM2/ΔFMΔFM2, brings about a posteriorization of caudal forebrain and midbrain into metencephalon, whereas the rostral forebrain development is unaffected (our unpublished data; Kurokawa et al, 2004a). The functions of OTX2 in head development have been relatively well analyzed (Acampora et al, 1995; Matsuo et al, 1995; Ang et al, 1996; Kimura et al, 2000; Puelles et al, 2003; Kurokawa et al, 2004a, 2004b; Vernay et al, 2005), but factors that function upstream or downstream of or cooperate with OTX2 at each stage and tissue are still poorly understood. The ultimate question of how Otx2 enhancer organization and gene cascades are established or conserved during the evolution of vertebrate head development also remains.
Efforts to determine the core elements essential to each enhancer activity have not been fully successful. The EP enhancer requires a 2.3 kb region; the FM enhancer could not be shortened beyond 1.4 kb without affecting its full activity, and the FM2 enhancer is 1.5 kb in length (Kurokawa et al, 2004a, 2004b). In contrast, the AN enhancer has been pinpointed to a 90 bp sequence. Potential SOX and HOX binding sequences exist in this 90 bp sequence, but mutation analysis indicated that these sequences are not essential to the activity. No other sequences for known DNA-binding factors exist in the 90 bp minimal AN enhancer sequence. One difficulty in identifying factors that bind to the 90 bp sequence is the limited size of the anterior neuroectoderm at the early somite stage.
In this study, to overcome the constraints imposed by size of the anterior neuroectoderm, we screened cultured cell lines and found that the AN enhancer is active in F9 cells. Using these cells, we identified the acetylated YY1 protein as a molecule that binds to the AN enhancer region. We also demonstrated that the AN enhancer activity is promoter-dependent, and this dependence is due to the necessity of the YY1-binding site in the promoter region. Both of these YY1 sites are essential for AN enhancer activity in mouse embryos at the presomite stage. Acetylated YY1 was found in the anterior head, in vivo, but not in the trunk, and the results of chromatin immunoprecipitation assay suggest that acetylated YY1 regulates Otx2 expression via the AN enhancer. Finally, the YY1 sites in the AN enhancer and promoter are highly conserved not only in tetrapods, but also in coelacanth and skate Otx2 loci.
Results
YY1 binds to AN enhancer in F9 cells
An EcoT22I/BglII 165 bp fragment (EB165) about −92 kb 5′ upstream of the Otx2 translation start site was previously determined by transgenic assay in mouse embryos to be necessary and sufficient for the Otx2 expression in anterior neuroectoderm at the presomite and early somite stages (Kurokawa et al, 2004a, 2004b). In our attempt to identify the factors that bind to the EB165 enhancer, to overcome the problem of the limited size of the anterior neuroectoderm, we screened cultured cell lines for the cells in which the AN enhancer is active. In this screen, we initially used tk or hsp68 promoter with the EB165; however, no AN-positive cell lines were obtained. The possibility of promoter dependence of the AN activity was then tested in anterior neuroectoderm at E7.75. The AN enhancer is active with the mouse 1.8 kb Otx2 gene endogenous promoter, as reported previously, but not with the hsp68 or tk promoters (Figure 1A and B). Rescreening of cell lines with the 1.8 kb promoter revealed that the AN enhancer is active in F9 cells; the enhancer was not active with the tk or hsp68 promoters in these cells. In the following analysis, NIH3T3 fibroblast cells served as a control; not only was the AN enhancer inactive, but the 1.8 kb promoter had no basal activity in the cells (Figure 1C). F9 cells are known to express a pan-neural marker, Sox2, as well as the epiblast markers Oct3/4 and Nanog (Kuroda et al, 2005). In addition, the F9 cells used in this study expressed anterior neural markers Six3 and Otx2, and weakly anterior mesendoderm markers Hnf3β, Lim1 and goosecoid (data not shown). The latter may explain the activity of the 1.8 kb region in F9 cells described below; this region harbors the enhancer for the expression in anterior mesendoderm (Figures 1A, 2D, and 4A) (Kimura et al, 2000).
Figure 1.
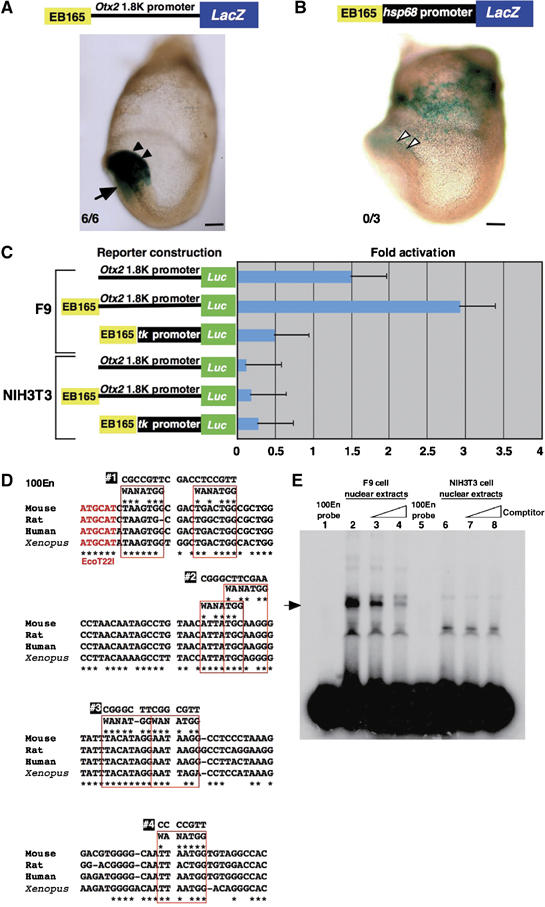
A protein–DNA complex is formed with F9 nuclear extracts and AN enhancer of which activity is promoter dependent. (A) The EB165 fragment directs the β-gal expression in the anterior neuroectoderm at presomite stage (E7.75) with the mouse Otx2 1.8 kb promoter (black arrowheads). Arrows indicate the expression in the anterior mesendoderm by the Otx2 1.8 kb promoter region (Kurokawa et al, 2004b). All six transgenic embryos generated were β-gal positive in anterior neuroectoderm. (B) The EB165 fragment is not able to drive the β-gal expression in the anterior neuroectoderm with the hsp68 promoter (white arrowheads). The β-gal expression in the extra-embryonic region is ectopic. Of the three transgenic embryos generated, none was β-gal positive in anterior neuroectoderm. Scale bars: 100 μm. (C) The EB165 fragment is active in F9 cells in a promoter-dependent manner, but inactive in NIH3T3 cells. Bars represent the mean standard errors of three independent experiments. (D) Approximately 100 bp sequences in the EB165 fragment are highly conserved among mouse, rat, human and Xenopus. The mouse DNA fragment was used as a probe in the EMSA and in the protein purification. Seven sites that partially match the YY1 core consensus sequences, WANATGG, are boxed in red. #1 to #4 indicate the sequences mutated for the competition experiments in the EMSA (Figure 2B and C). (E) EMSA using F9 nuclear extracts exhibits a protein–DNA complex (arrow) with the 100En probe, but does not do so using NIH3T3 nuclear extracts. The specificity of the binding is demonstrated by competition with an increasing amount of unlabeled 100En, from 10- to 100-fold excess.
Figure 2.
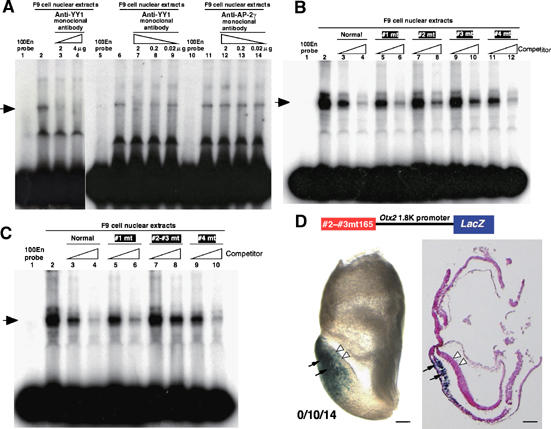
YY1 binding to AN enhancer is essential to its activity in anterior neuroectoderm. (A) Anti-YY1 monoclonal antibody inhibits the DNA–protein complex formation (lanes 3, 4 and 7–9), whereas a nonspecific antibody against AP-2γ has no effect (lanes 12–14). F9 nuclear extracts preincubated with anti-YY1 monoclonal antibody was subjected to EMSA with 100En probe labeled with [32P]-dCTP. (B) Competition experiments with four 100En probes mutated at potential YY1 binding sequences; see Figure 1D for mutated sequences in #1–#4 100En probes. The #1 or #4 100En probe decreases the signal of the DNA–protein complex as the normal 100En probe does (lanes 3–6, 11 and 12). However, with the #2 or #3 100En probe as a competitor, some signals remain (lane 7–10). (C) Competition experiments with the 100En probe with both #2 and #3 mutations. The probe lost the competitive activity (lanes 7 and 8). (D) The EB165 AN enhancer with both #2 and #3 mutations loses its enhancer activity in the anterior neuroectoderm at presomite stage (white arrowheads). Among 14 transgenic embryos generated, 10 exhibited β-gal expression in anterior mesendoderm by the 1.8 kb promoter region (Figure 1A); none of these exhibited β-gal expression in anterior neuroectoderm. Scale bars, 100 μm.
Figure 4.
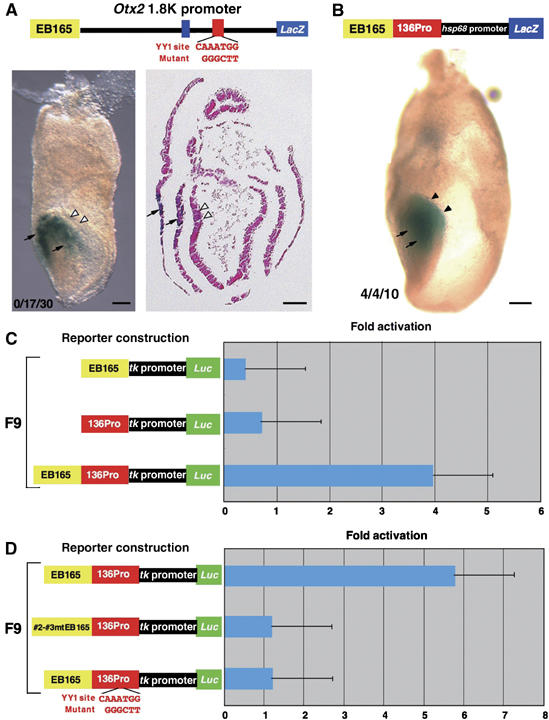
YY1 binding to both the AN enhancer and promoter is required for activity. (A) The mutation in the YY1 site of the 1.8 kb promoter region abolishes the EB165 enhancer activity in anterior neuroectoderm at presomite stage (white arrowheads); however, enhancer activities of the 1.8 kb promoter region in the anterior mesendoderm, visceral endoderm and cephalic mesenchyme are unaffected (arrows; data not shown). cf. Figure 1A. Among 30 transgenic embryos generated, seventeen exhibited β-gal expression in anterior mesendoderm by the 1.8 kb promoter region; none exhibited β-gal expression in anterior neuroectoderm. (B) 136Pro endows the enhancer activity in anterior neuroectoderm (black arrowheads) to EB165 enhancer and hsp68 promoter (cf. Figure 1B). The 136Pro fragment also harbors the enhancer activity for the expression in anterior mesendoderm (arrows). Among 10 transgenic embryos generated, four exhibited β-gal expression in anterior mesendoderm by the 1.8 kb promoter region; these four embryos also exhibited β-gal expression in anterior neuroectoderm. Scale bars, 100 μm. (C) The 136Pro fragment also endows the enhancer activity to the EB enhancer and tk promoter in F9 cells; the fragment itself does not have enhancer activity in F9 cells. (D) The activity is lost by the mutation of the YY1-binding site in either EB165 enhancer or 136Pro alone in F9 cells.
In the EB165 enhancer, a sequence of about 90 bp is particularly well conserved in mouse, rat, human and Xenopus Otx2 loci (Figure 1D), and these were previously confirmed as essential for the AN activity in vivo (Kurokawa et al, 2004b). An electrophoretic mobility shift assay (EMSA) was conducted using the 100 bp DNA fragment covering these sequences as a probe (100En probe). The shift band by DNA–protein complex formation was observed with the nuclear extracts of the F9 cells, but not with those of NIH3T3 cells (Figure 1E, lanes 2 and 6). Competition analysis with increasing amounts of unlabelled probe decreased the signal of the shift band (Figure 1E, lanes 3 and 4), indicating that the binding activity is specific to the 100En probe.
To identify proteins that bind to the 100En probe, we employed one-step affinity purification. F9 nuclear extracts were applied to the streptavidin magnetic particles to which biotin-labeled 100 En probe was immobilized and the DNA–protein complexes were collected with a magnet (Kaneoka et al, 1991). The eluate fraction from the particles exhibited a strong signal, indicating complex formation with the 100En probe in EMSA. Comparing the SDS–PAGE gel pattern of the eluate fraction with those of the crude F9 nuclear extracts and the supernatant fraction that was not bound to the particles, three protein bands were identified to be unique to the eluate fraction. These were HSP-70, EF-Tu and YY1, a GLI–Krüppel zinc-finger transcription factor (Supplementary Figure 1).
YY1 binding to the EB165 region is essential to the AN enhancer activity
To confirm that YY1 is indeed a component of the DNA–protein complex in EMSA (Figure 1E), the effect of the anti-YY1 monoclonal antibody on EMSA was tested. The DNA–protein complex was not supershifted by the addition of specific antibody; the anti-YY1 antibody diminished the complex under the reaction condition used (Figure 2A, lanes 3 and 4). With the reduction in the amount of antibody the intensity of the shift band was restored (Figure 2A, lanes 7–9), whereas a nonspecific monoclonal antibody had no effect on the DNA–protein complex (Figure 2A, lanes 12–14). These suggest that the YY1 protein is indeed the functional component of the DNA–protein complex in EMSA.
The core consensus sequence of YY1 binding is WANATGG (Shi et al, 1997). The 100En region of the EB165 enhancer does not contain this core sequence; however, there are seven partial-match sites to the core sequence (Figure 1D, red box). To examine whether YY1 protein binds to these sites, a transverse mutation was introduced into them as indicated in Figure 1D (#1–#4), and each mutant 100En was tested as an unlabeled competitor in the EMSA. The #1 or #4 mutant competitor decreased the signal of DNA–protein complex in the EMSA as the normal competitor did; however, each #2 or #3 mutant decreased the signal only partially (Figure 2B). This raised the question of whether YY1 might bind the 30 bp region over these two mutations. Indeed, the 100En fragment in which both #2 and #3 are mutated lost the activity of the competitor (Figure 2C, lanes 7 and 8).
To confirm the significance of this 30 bp region for the AN enhancer activity, the in vivo activity of the EB165 fragment with the mutation at both the #2 and #3 sites was examined by transgenic assay; the mutation abolished AN activity (Figure 2D).
YY1 binding to the promoter region is also essential for the AN enhancer activity
The AN enhancer exhibited promoter dependency as described above (Figure 1A, B and C). In the 1.8 kb promoter region two sites are highly conserved in mouse, chick and Xenopus (Figure 3A). One of these, −863 to −825 bp upstream of the mouse Otx2 translation start site (Figure 3A, blue box) harbors the enhancer directing the expression of Otx2 in cephalic neural crest cells that we previously identified (Kimura et al, 1997). The other is located −603 and −468 bp upstream (Figure 3A, red box). In this 136 bp sequence, there exists a consensus YY1-binding site, WANATGG, (Figure 3B, red nucleotides). The mouse Otx2 gene is reported to possess three transcription start sites, the usage of which appears to differ by cell type and developmental stage (Fossat et al, 2005). The most proximal one that is proposed to be most active during early embryonic development locates −207 bp 5′ upstream of the translation initiation site (Figure 3A). The other two transcriptional start sites exist 5′ farther upstream than 1.8 kb.
Figure 3.
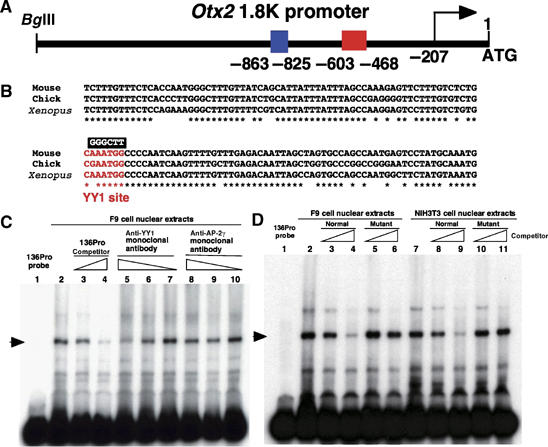
YY1 also binds to the WANATGG sequence in the 1.8 kb promoter region. (A) Schematic representation of the mouse 1.8 kb promoter region. In this region there are two sites highly conserved among mouse, chick and Xenopus at −863∼−825 (blue box) and −603∼−468 (red box) 5′ upstream of the translation start site, ATG. The site at −863∼−825 harbors the enhancer directing the Otx2 expression in cephalic mesenchyme (Kimura et al, 1997). There are three Otx2 transcription starts and the most proximal one locates at −207 bp 5′ upstream of the translational start site (Fossat et al, 2005). (B) Alignments of 136 bases in the red box region in panel A among mouse, chick and Xenopus. This region was used as a probe, 136Pro, in the EMSA. The red letters indicate the core consensus sequences for the YY1 transcription factor. GGGCTT indicates the sequence in a mutated 136Pro probe for the competition experiment in EMSA (Figure 3D). (C) The DNA–protein complex formation between F9 nuclear extracts and labeled 136Pro probe is in competition with unlabeled 136Pro (lanes 3 and 4) and anti-YY1 monoclonal antibody (lanes 5 and 6), whereas a nonspecific antibody against AP-2γ has no effect (lanes 8–10). (D) Both F9 and NIH3T3 nuclear extracts form protein–DNA complexes with 136Pro (lanes 2–4, 7–9). The 136Pro mutated at the WANATGG sequence does not compete for the complex formation with either F9 nuclear extracts (lanes 5 and 6) or NIH3T3 nuclear extracts (lanes 10 and 11).
To confirm YY1-binding to the WANATGG sequence, EMSA was performed using the 136 bp fragment (136Pro) probe and F9 nuclear extract. The anti-YY1 monoclonal antibody reduced the signal of the DNA–protein complex (Figure 3C, lane 5), and the signal was restored by a decrease in amount of the antibody (Figure 3C, lanes 6 and 7). A nonspecific monoclonal antibody had no effect on DNA–protein complex formation (Figure 3C, lanes 8–10); complex formation was also unaffected by a mutant 136Pro fragment, in which a transverse mutation was introduced into the YY1-binding site (Figure 3D, lanes 5 and 6). These data suggest that YY1 protein also binds to the 136Pro region of the 1.8 kb Otx2 gene endogenous promoter.
To confirm the role of the YY1-binding site in the 136Pro region for the Otx2 expression in anterior neuroectoderm, transgenic analysis was conducted with the wild-type AN enhancer and the 1.8 kb promoter in which the YY1 site in 136Pro was mutated. The transverse mutation in the YY1-binding site abolished β-gal expression in the AN (Figure 4A; cf. Figure 1A). The 1.8 kb region harbors the enhancer activities for the expression in anterior visceral endoderm, anterior mesendoderm and cephalic neural crest cells (Kimura et al, 1997; Kimura et al, 2000). The transverse mutation in the YY1 site did not affect these activities (Figure 4A; data not shown).
Next we assessed whether the YY1 site in the 136Pro region is sufficient to explain the 1.8 kb promoter dependence of the AN enhancer activity. The EB165 AN enhancer in combination with tk (EB165tk) or hsp68 (EB165hsp) promoter is inactive not only in vivo, as described above, but also in vitro (Figure 1B, C and 4C). 136Pro activated inactive EB165tk and EB165hsp both in vivo (Figure 4B) and in vitro (Figure 4C), whereas it did not activate the tk or hsp promoters in these assays (Figure 4C; data not shown). Furthermore, mutations of either the #2-#3 YY1 site in EB165 or the YY1 site in 136Pro abolished luciferase expression in F9 cells (Figure 4D). Together, these findings indicate that the simultaneous YY1 binding to both YY1 sites in the 100En region of the EB165 AN enhancer and in the 136Pro region of the 1.8 kb promoter is essential to the AN activity.
YY1 is known to promote transcription not only in the presence of TBP (TATA-binding protein), but also in its absence; YY1 is able to recruit TIFs (transcriptional initiation factors) by itself (Usheva and Shenk, 1994). However, the EB165/130Pro was not active in the absence of hsp68 or tk promoter either in E7.75 AN or F9 cells (Supplementary Figure 2A and B), indicating that YY1 binding to 136Pro is unable to assemble TIFs by itself. Therefore, in the mouse endogenous Otx2 locus, a sequence must exist between the transcription start site at −207 bp and the 136Pro site at −468 bp 5′ upstream (Supplementary Figure 2C) that recruits TIFs. A deletion analysis indicated that the region between −300 and −208 bp 5′ upstream is essential for EB165/136Pro activity in E7.75 AN (Supplementary Figure 2C). The region has a TATA-like sequence (Supplementary Figure 2D).
Only acetylated YY1 binds to YY1 sites in the AN enhancer
Apparently controversial is the fact that NIH3T3 cells express YY1 (Austen et al, 1997), although at a level much lower than that of F9 cells (Supplementary Figure 3A). NIH3T3 nuclear extracts formed the DNA–protein complex with the 136Pro probe (Figure 3D, lanes 7–11), and the anti-YY1 monoclonal antibody blocked the interaction (data not shown). Thus, the question remains as to why YY1 protein in NIH3T3 nuclear extracts was unable to activate EB165 AN enhancer (Figure 1C) or bind to 100 En probe (Figure 1E).
The DNA binding activity of YY1 protein is reported to be affected by acetylation (Yao et al, 2001). There was acetylated YY1 in F9 nuclear extracts, but acetylation of YY1 in NIH3T3 nuclear extracts was scant (Supplementary Figure 3B). To test the possibility that acetylation explains the apparent discrepancy surrounding YY1, the effect of anti-acetylated-lysine antibody on EMSA was examined. The antibody inhibited the complex formation between F9 nuclear extracts and 100En probe (Figure 5A, lane 3), whereas it only moderately decreased complex formation between the nuclear extracts and 136Pro probe (Figure 5A, lane 6). Furthermore, the treatment of F9 nuclear extracts with histone deacetylase completely abolished complex formation with the 100En probe, whereas the treatment did not affect complex formation with the 136Pro probe (Figure 5B, lanes 3 and 7).
Figure 5.
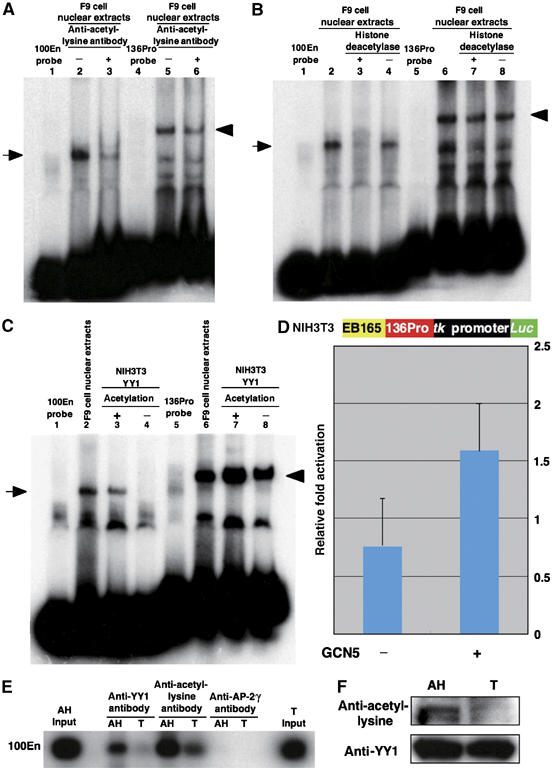
Acetylated YY1, but not unmodifiedYY1, binds to 100En. (A) The anti-acetylated-lysine monoclonal antibody significantly reduces the DNA–protein complex formation between F9 nuclear extracts and the 100En probe (arrow, lane 3), but only slightly that between the extracts and 136Pro probe (arrowhead, lane 6). (B) Deacetylation of F9 nuclear extracts with histone deacetylase abolishes complex formation with the 100En probe (arrow, lane 3), but exhibits no effect on that with the 136Pro probe (arrowhead, lane 7). (C) YY1 protein purified from NIH3T3 nuclear extracts is unable to bind to the 100En probe (arrow, lane 4), but binds to the 136Pro probe (arrowhead, lane 8). However, the binding to the 100En probe occurs when the YY1 protein from NIH3T3 nuclear extracts is acetylated with yeast GCN5 (lane 3); the acetylation increases the binding to the 136Pro probe (lanes 7 and 8). (D) YY1 acetylation with human Gcn5 activates the EB165 enhancer with 136Pro in NIH3T3 cells. The tk-promoter/luciferase reporter with the EB165 and 136Pro fragments was transfected to NIH3T3 cells together with the Yy1 expression vector either with or without the human Gcn5 expression vector. (E) ChIP assay with anti-YY1 or anti-acetylated-lysine antibody of anterior head (AH) or trunk (T) at E7.75; AH is the AN enhancer-positive anterior head and T is the AN enhancer-negative trunk (see Materials and methods for details). The AP-2γ antibody, a negative control, did not precipitate the 100En in either AN or T. The 100En amplification with the AH and T input is also given. (F) The YY1 protein is acetylated in the anterior head (AH) but not in the trunk (T) (upper panel). The amounts of YY1 protein immunoprecipitated with anti-YY1 antibody are nearly equal between the anterior head and the trunk (lower panel).
We then attempted to acetylate YY1 protein purified from the NIH3T3 nuclear extracts. This assay, which incorporated labeled acetyl coenzyme A into YY1 protein, demonstrated that yeast GCN5 can acetylate YY1 protein (data not shown). YY1 protein thus acetylated was able to bind to the YY1 binding sequences in the 100En probe (Figure 5C, lane 3). Unmodified YY1 protein was unable to bind to this probe, although it was able to bind to 136Pro probe (Figure 5C, lanes 4 and 8). Although YY1 binding to the adeno-associated virus P5 promoter is reduced by the acetylation (Yao et al, 2001), acetylated YY1 protein rather efficiently bound to the 136Pro probe (Figure 5C, lanes 7 and 8). The same results were also obtained with PCAF, a mammalian histone acetyltransferase (Supplementary Figure 3C). We also examined whether the acetylation of YY1 protein can activate transcriptional machinery consisting of the EB165 AN enhancer, 136Pro and tk promoter in NIH3T3 cells. Cotransfection of human Gcn5 acetyltransferase gene with the EB165-136Pro/tk-luciferase construct induced luciferase expression (Figure 5D).
One major remaining question is whether the AN enhancer is regulated by acetylated YY1 in vivo. To assess this, a chromatin immunoprecipitation (ChIP) assay was conducted using a transgenic line that harbors the LacZ gene under the AN enhancer (Kurokawa et al, 2004b). Presomitic embryos were dissected into β-gal-positive anterior head and β-gal-negative trunk sections; β-gal-negative non-neuroectodermal tissues were removed as much as possible in the head. In the ChIP assay using an anti-YY1 antibody, the 100En region precipitated in the anterior head fraction but not in the trunk. The anti-acetylated-lysine antibody precipitated the 100En region in the anterior head fraction at a level of about threefold that of the trunk (Figure 5E). The acetylated YY1 protein was consistently found in the anterior head fraction (Figure 5F). These results strongly suggest that, in vivo, Otx2 expression in the anterior neuroectoderm under the control of the AN enhancer is regulated by the acetylation of the YY1 protein.
A recent study by our group indicated that the AN enhancer is conserved not only among tetrapod Otx2 cognates (Kurokawa et al, 2004b), but also in coelacanth and skate Otx2 cognates; the AN regions in the Otx2 loci of these animals exhibited activity in mouse AN (Kurokawa et al, 2006). The conservation of YY1 sites was examined in their AN enhancers and promoter regions (Figure 6A and B). As expected, YY1-binding sequences are highly conserved in both regions of coelacanth and skate Otx2 loci; moreover, they bound YY1 from F9 cells (Figure 6C and D, Supplementary Figure 4).
Figure 6.
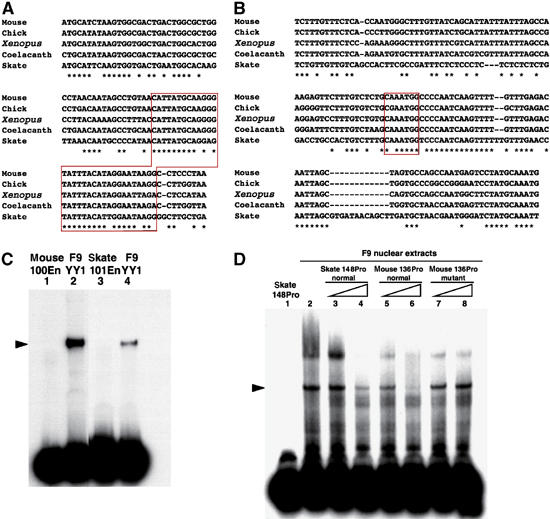
YY1 sites in the AN enhancer and promoter are conserved among tetrapods, coelacanth and skate. (A) Alignment of 100En domain sequences in the EB165 enhancer among mouse, chick, Xenopus, coelacanth and skate Otx2 loci; in skate it is 101 bp (101En). The 30 bp sequences around #2 and #3 mutations in Figure 1D, which are recognized by the acetylated YY1 protein, are indicated in a red box. (B) Alignment of the 136Pro domain sequences in the promoter region among mouse, chick, Xenopus, coelacanth and skate Otx2 loci; in skate it is 148 bp (148Pro). The YY1 core consensus sequence is conserved among these animals (red box). (C) YY1 from F9 cells binds to skate 101En (lane 4), although less efficiently than to mouse 100En (lane 2). (D) Nuclear extracts of F9 cells form a DNA–protein complex with skate 148Pro (lanes 2–4) that is competed with mouse 136Pro (lanes 5 and 6), but not with its mutant version (lanes 7 and 8; see Figure 3B).
Discussion
This study demonstrated that mouse Otx2 expression in anterior neuroectoderm is regulated by acetylated YY1, which binds to a 30 bp site in the EB165 enhancer about 92 kb upstream and to the consensus YY1-binding sequence in the promoter region about 530 bp upstream of the translation start site. The sequence in the promoter region contains the core consensus sequence of YY1 binding, whereas the site in the enhancer has sequences that only partially match the core sequence. Only acetylated YY1 can bind to the site in the enhancer, whereas both acetylated and non-acetylated YY1 can bind to the site in the promoter region. YY1 binding to both of these sites is required for the Otx2 expression, which is essential for development of the anterior neuroectoderm.
Of the enhancers responsible for Otx2 expression in various contexts (Kimura et al, 1997, 2000; Kurokawa et al, 2004a, 2004b), the YY1-binding site in the promoter region is required specifically for the AN enhancer activity. The AN enhancer region is an essential component of the EP enhancer, but the latter is active with the hsp promoter (Kurokawa et al, 2004b). The Otx2 expression in the anterior neuroectoderm is regulated by the FM enhancer 75 kb 5′ upstream at stages later than E8.5. This enhancer is also active with hsp promoter (Kurokawa et al, 2004a). The AVE, AME and CNC enhancers locate in the promoter region (Kimura et al, 1997; Kimura et al, 2000). A mutation in the YY1 site did not affect these enhancer activities of the 1.8 kb promoter region (Figure 4A, data not shown).
The AN enhancer is conserved not only among tetrapod Otx2 cognates (Kurokawa et al, 2004b), but also in coelacanth and skate Otx2 cognates (Kurokawa et al, 2006). In addition, YY1 sites are conserved in both the AN enhancer and promoter regions of both coelacanth and skate Otx2 cognates. Moreover, YY1 from F9 cells binds to the skate enhancer and promoter. These findings strongly suggest that YY1 regulation of the AN enhancer was established concurrently with the AN enhancer in an ancestral gnathostome and has been conserved, together with the enhancer, in taxa up to and including mammal.
YY1 (Yin Yang 1) is a transcription factor with a GLI-Krüppel type zinc-finger motif highly related to pleiohomeotic (Shi et al, 1991; Flanagan et al, 1992; Brown et al, 1998). It is a multifunctional factor that modulates the expression of a wide variety of genes acting as a transcriptional activator or repressor in a promoter context-dependent manner (Shi et al, 1997; Thomas and Seto, 1999). In addition, it can cause DNA bending and is involved in chromatin remodeling. A wide variety of proteins are known to associate with YY1; they may differentially regulate YY1 activities on each target gene in each cell. Of particular interest for this study are the associations with histone acetyltransferases and histone deacetylases. YY1 can be acetylated by p300 and PCAF (p300-CBP-associated factor); acetylation alters the DNA-binding and transcriptional activity of the YY1 protein (Yao et al, 2001). Little is known, however, about the precise role of YY1 in vertebrate embryonic development, nor has anything been demonstrated about the in vivo significance of its acetylation. This study is the first to demonstrate that the regulation of YY1 activity by acetylation plays a decisive role in Otx2 expression crucial to the development of anterior neuroectoderm. Acetylated, but not unmodified, YY1 can recognize the sequence in the enhancer, whereas both can bind to the sequence in the promoter region. DNA binding sequences for YY1 binding appear to be variable by cofactors and modification (Shi et al, 1997; Thomas and Seto, 1999). Acetylation of YY1 in the zinc-finger domain was shown to diminish the binding efficiency to adeno-associated virus P5 promoter (Yao et al, 2001). The precise sequence in the AN enhancer for the unique recognition by the acetylated YY1 remains for future studies.
YY1 transcripts are present ubiquitously during mouse development, and the null mutant is lethal at the peri-implantation stage, precluding the examination of its role in the development of anterior neuroectoderm (Donohoe et al, 1999). However, studies in Xenopus support our result. YY1 overexpression induces neural tissue (Satijn et al, 2001). Moreover, YY1 knockdown by antisense morpholino oligonucleotide reduces anterior neuroectoderm with the decrease in Otx2 expression (Kwon and Chung, 2003). In Xenopus Otx2 locus, YY1-binding sites are conserved in both the AN enhancer and promoter regions. The Xenopus system should be useful to further clarify the role of the YY1 regulation in Otx2 expression and anterior neuroectoderm development.
Unfortunately, no antibody against acetylated YY1 is available to allow us to demonstrate that acetylated YY1 localizes at the AN or to examine how it is specifically acetylated there. We have, however, shown that acetylated YY1 is found in the anterior head but not in the trunk. And our ChIP assay has demonstrated that YY1 associates with the AN enhancer in anterior head, but not in the trunk.
Mouse mutants of Pcaf, Gcn5l2, p300 or cbp have been reported (Yao et al, 1998; Xu et al, 2000; Yamauchi et al, 2000), but they exhibit defects at multiple sites and tell little about the roles of these genes in development of anterior neuroectoderm, Otx2 expression or YY1 acetylation. The zinc-finger domain of YY1 is acetylated by PCAF (Yao et al, 2001). Pcaf is not expressed, but Gcn5l2 is strongly expressed in mouse embryos at the headfold stage (Xu et al, 2000; Yamauchi et al, 2000). It has been suggested that GCN5l2 is an acetyltransferase with substrate specificity very similar to that of PCAF (Xu et al, 1998). Notably, Otx2 expression is absent in an ectoderm-like structure developing in the anterior portion of the Gcn5l2 mutant (Xu et al, 2000). It would be interesting to examine a Gcn5l2 conditional knockout with Cre under the control of the AN enhancer.
Also worth noting is the fact that YY1 is necessary but not sufficient to drive the AN enhancer. YY1 binding to the 136Pro promoter region is unable to assemble TIFs by itself; it is dependent on a domain that has TATA-like sequence. A number of functions of YY1 have been considered to require coactivators in the TATA-dependent transcriptional activation (Shi et al, 1997; Thomas and Seto, 1999). Moreover, besides the YY1-binding site, several sequences are also conserved in the 130Pro region among skate, coelacanth and tetrapods, suggesting that there are several other factors that bind to these sequences (Figure 6B). Acetylated YY1 binding is essential but also not sufficient to drive the EB165 AN enhancer. The deeply conserved 90 bp sequence is required for its activity; multiple factors must also bind to this sequence (Figure 6A; Kurokawa et al, 2004b). In contrast to the transgenic assay in mouse embryos, the response to the activation assay in the cell culture system was moderate. Apparently the in vitro system must be oversimplified; other developmentally regulated transcriptional factors must not be present sufficiently. The identification of the factors is asked to elucidate the upstream cascade of the OTX2 function in anterior neuroectoderm.
Materials and methods
Preparation of nuclear extracts
F9 teratocarcinoma cells and NIH3T3 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 1% penicillin/streptomycin. When the cells were 80∼90% confluent, they were harvested to prepare cytoplasmic and nuclear extracts using NE-PER Nuclear and Cytoplasmic Extraction Reagents (PIERCE). A Protein Assay Rapid kit (Wako Pure Chemical Industries, Osaka) was utilized to calculate an approximate concentration of protein in each fraction.
Electrophoretic mobility shift assay
The 100En probe was prepared by the PCR amplification with oligonucleotides (5′-TCAAGCTTTCTAAGTGGCGACTGACTGG-3′) and (5′-GTAAGCTTACACCATTAATTGCCCCACG-3′) with the HindIII recognition sequence at their 5′ ends. The PCR product was digested with HindIII and purified by agarose gel electrophoresis. The 136Pro probe was similarly prepared with oligonucleotides (5′-TTCAAGCTTCTTTGTTTCTCACCAATGGG-3′) and (5′-AGTAAGCTT CATTTGCATAGGACTCATTGGC-3′). The purified probes were labeled with [32P]-dCTP by the Klenow fragment of DNA polymerase I (New England Biolabs). For EMSA analysis, nuclear protein extracts were incubated in the reaction mixtures as described (Sakamoto et al, 1997). After incubation, the reaction mixtures were separated on 3.5% non-denaturing polyacrylamide gels. For blocking experiments with antibodies, anti-YY1 (SC-7341X, Santa Cruz) or anti-acetylated-lysine (clone 4G12, UPSTATE) antibody was preincubated with nuclear extracts for 15 min at 4°C, and then incubated with 100En or 136Pro probe at 13°C for 30 min. For the deacetylation, nuclear extracts were incubated with Rat Liver Histone Deacetylase (CALBIOCHEM) at 30°C for 30 min in advance of the EMSA. The acetylation of YY1 protein purified from NIH3T3 nuclear extracts was conducted with Saccharomyces cerevisiae GCN5 protein and human PCAF (UPSTATE) as described (Kuo et al, 1996).
Partial purification of YY1 protein from F9 and NIH3T3 cells
F9 and NIH3T3 nuclear extracts were prepared as described above. By taking advantage of the long consecutive histidine stretch in the YY1 protein, the protein was isolated from NIH3T3 nuclear extracts using Ni-NTA magnetic agarose beads (Qiagen) (Rezai-Zadeh et al, 2003).
Construction of reporter transgenes
The VEcis-lacZ (Kurokawa et al, 2004a, 2004b) and the ASShsplacZpA (Nishizaki et al, 2001) vectors were the sources used to construct reporter transgenes for in vivo assay with 1.8 kb and hsp promoters, respectively. The pGL3-Basic luciferase reporter vector (Promega) was used to generate reporter constructs for the in vitro assay. Each YY1-binding site in EB165 enhancer fragment or 1.8 kb promoter was mutated by transverse substitution using the PCR-based overlap extension method (Kucharczuk et al, 1999).
Production of transgenic embryos
Vectors containing transgenes were digested with appropriate restriction enzymes to remove vector-derived sequences, electrophoresed on agarose gels and purified from the gels using a QIAquick Gel Extraction kit (Qiagen). Transgene DNAs (1–2 ng/μl) were injected into the pronuclei of fertilized CD-1 mouse zygotes as described (Kurokawa et al, 2004b). Transgenic embryos were determined by PCR with oligonucleotide primers specific to the LacZ gene (Sasaki et al, 1997), and β-gal expression was detected as described (Kimura et al, 1997).
Luciferase reporter assays
F9 or NIH3T3 cells were incubated in 24-well tissue culture plates for 24 h. The cells were then transfected with each reporter construct by Lipofectamin 2000 (Invitrogen) according to the manufacturer's protocol. To determine the effects of YY1-acetylation in NIH3T3 cells, the reporter construct was cointroduced into the cells with the full-length Yy1 cDNA and the full-length human Gcn5 cDNA in the pCMV expression vector (Open Biosystems). Cell extracts were prepared 24 h after the transfection. The luciferase activity was determined as described: LacZ gene in the pCS2 vector was cotransfected, and the luciferase activity was normalized with β-galactosidase activity (Yamamoto et al, 2005). Each assay was performed in triplicate and repeated at least three times.
Chromatin immunoprecipitation
A transgenic line harboring the LacZ gene under the AN enhancer was established previously (Kurokawa et al, 2004b). About 70 E7.8 embryos were dissected into the β-gal-positive anterior head and the β-gal-negative trunk; β-gal-negative non-neuroectodermal tissues were minimal in the former but not removed in the latter. The tissues were treated for 10 min with 1% formaldehyde at 37°C, and the ChIP assay was conducted according to the protocol by Upstate Biotechnology (Lake Placid, NY). The fixed tissues were sonicated in SDS lysis buffer (50 mM Tris–HCl, pH 8.1, 10 mM EDTA and 1% SDS) with Handy Sonic (TOMY SEIKO Co., Tokyo) to generate DNA fragmentation at the size range from 200 to 1000 bp. The lysates were incubated with 10 μg of each antibody at 4°C for overnight; complexes were collected with recombinant protein A agarose beads. After washing the precipitates, eluting the complexes from the beads and reverse crosslinking from formaldehyde, DNA fragments were purified with phenol/chloroform extraction. Amplification by PCR was performed with Taq DNA polymerase (Promega) using oligonucleotides specific to 100En (5′-TCTAAGTGGCGACTGACTGG-3′, 5′-CTTTATGGAAGAGAGGACCG-3′). PCR products were electrophoresed on 2% agarose gel and then transferred onto the nylon membrane for Southern hybridization using a 100En probe.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 1 Legend
Supplementary Figure 2
Supplementary Figure 2 Legend
Supplementary Figure 3
Supplementary Figure 3 Legend
Supplementary Figure 4
Supplementary Figure 4 Legend
Supplementary Materials and Methods
Acknowledgments
We thank Dr T Kuno for providing us the yeast Gcn5 cDNA plasmid and Dr K Kokura for critical discussion. We are also grateful to the Laboratory for Animal Resource and Genetic Engineering for the generation and housing of transgenic mice. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brulet P (1995) Forebrain and midbrain regions are deleted in Otx2−/− mutants due to a defective anterior neuroectoderm specification during gastrulation. Development 121: 3279–3290 [DOI] [PubMed] [Google Scholar]
- Ang SL, Jin O, Rhinn M, Daigle N, Stevenson L, Rossant J (1996) A targeted mouse Otx2 mutation leads to severe defects in gastrulation and formation of axial mesoderm and to deletion of rostral brain. Development 122: 243–252 [DOI] [PubMed] [Google Scholar]
- Austen M, Luscher B, Luscher-Firzlaff JM (1997) Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J Biol Chem 272: 1709–1717 [DOI] [PubMed] [Google Scholar]
- Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA (1998) The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell 1: 1057–1064 [DOI] [PubMed] [Google Scholar]
- Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y (1999) Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol 19: 7237–7244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JR, Becker KG, Ennist DL, Gleason SL, Driggers PH, Levi BZ, Appella E, Ozato K (1992) Cloning of a negative transcription factor that binds to the upstream conserved region of Moloney murine leukemia virus. Mol Cell Biol 12: 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossat N, Courtois V, Chatelain G, Brun G, Lamonerie T (2005) Alternative usage of promoters during mouse development. Dev Dyn 233: 154–160 [DOI] [PubMed] [Google Scholar]
- Kaneoka H, Lee DR, Hsu KC, Sharp GC, Hoffman RW (1991) Solid-phase direct DNA sequencing of allele-specific polymerase chain reaction-amplified HLA-DR genes. Biotechniques 10: 30–34 [PubMed] [Google Scholar]
- Kimura C, Takeda N, Suzuki M, Oshimura M, Aizawa S, Matsuo I (1997) Cis-acting elements conserved between mouse and pufferfish Otx2 genes govern the expression in mesencephalic neural crest cells. Development 124: 3929–3941 [DOI] [PubMed] [Google Scholar]
- Kimura C, Yoshinaga K, Tian E, Suzuki M, Aizawa S, Matsuo I (2000) Visceral endoderm mediates forebrain development by suppressing posteriorizing signals. Dev Biol 225: 304–321 [DOI] [PubMed] [Google Scholar]
- Kimura J, Suda Y, Kurokawa D, Hossain ZM, Nakamura M, Takahashi M, Hara A, Aizawa S (2005) Emx2 and Pax6 function in cooperation with Otx2 and Otx1 to develop caudal forebrain primordium that includes future archipallium. J Neurosci 25: 5097–5108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharczuk KL, Love CM, Dougherty NM, Goldhamer DJ (1999) Fine-scale transgenic mapping of the MyoD core enhancer: MyoD is regulated by distinct but overlapping mechanisms in myotomal and non-myotomal muscle lineages. Development 126: 1957–1965 [DOI] [PubMed] [Google Scholar]
- Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD (1996) Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383: 269–272 [DOI] [PubMed] [Google Scholar]
- Kuroda T, Tada M, Kubota H, Kimura H, Hatano SY, Suemori H, Nakatsuji N, Tada T (2005) Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol Cell Biol 25: 2475–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa D, Kiyonari H, Nakayama R, Kimura-Yoshida C, Matsuo I, Aizawa S (2004a) Regulation of Otx2 expression and its functions in mouse forebrain and midbrain. Development 131: 3319–3331 [DOI] [PubMed] [Google Scholar]
- Kurokawa D, Sakurai Y, Inoue A, Nakayama R, Takasaki N, Suda Y, Miyake T, Amemiya C, Aizawa S (2006) Evolutionary constraint on Otx2-neuroectoderm enhancers; deep conservation from skate to mouse and unique divergence in teleost. Proc Natl Acad Sci USA 103: 19350–19355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa D, Takasaki N, Kiyonari H, Nakayama R, Kimura-Yoshida C, Matsuo I, Aizawa S (2004b) Regulation of Otx2 expression and its functions in mouse epiblast and anterior neuroectoderm. Development 131: 3307–3317 [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Chung HM (2003) Yin Yang 1, a vertebrate polycomb group gene, regulates antero-posterior neural patterning. Biochem Biophys Res Commun 306: 1008–1013 [DOI] [PubMed] [Google Scholar]
- Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S (1995) Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev 9: 2646–2658 [DOI] [PubMed] [Google Scholar]
- Nishizaki Y, Shimazu K, Kondoh H, Sasaki H (2001) Identification of essential sequence motifs in the node/notochord enhancer of Foxa2 (Hnf3β) gene that are conserved across vertebrate species. Mech Dev 102: 57–66 [DOI] [PubMed] [Google Scholar]
- Puelles E, Acampora D, Lacroix E, Signore M, Annino A, Tuorto F, Filosa S, Corte G, Wurst W, Ang SL, Simeone A (2003) Otx dose-dependent integrated control of antero-posterior and dorso-ventral patterning of midbrain. Nat Neurosci 6: 453–460 [DOI] [PubMed] [Google Scholar]
- Rezai-Zadeh N, Zhang X, Namour F, Fejer G, Wen YD, Yao YL, Gyory I, Wright K, Seto E (2003) Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev 17: 1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto N, Akasaka K, Mitsunaga-Nakatsubo K, Takata K, Nishitani T, Shimada H (1997) Two isoforms of orthodenticle-related proteins (HpOtx) bind to the enhancer element of sea urchin arylsulfatase gene. Dev Biol 181: 284–295 [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hui C, Nakafuku M, Kondoh H (1997) A binding site for Gli proteins is essential for HNF-3β floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development 124: 1313–1322 [DOI] [PubMed] [Google Scholar]
- Satijn DP, Hamer KM, den Blaauwen J, Otte AP (2001) The polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol Cell Biol 21: 1360–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lee JS, Galvin KM (1997) Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta 1332: F49–F66 [DOI] [PubMed] [Google Scholar]
- Shi Y, Seto E, Chang LS, Shenk T (1991) Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67: 377–388 [DOI] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E (1992) Nested expression domains of four homeobox genes in developing rostral brain. Nature 358: 687–690 [DOI] [PubMed] [Google Scholar]
- Thomas MJ, Seto E (1999) Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene 236: 197–208 [DOI] [PubMed] [Google Scholar]
- Usheva A, Shenk T (1994) TATA-binding protein-independent initiation: YY1, TFIIB, and RNA polymerase II direct basal transcription on supercoiled template DNA. Cell 76: 1115–1121 [DOI] [PubMed] [Google Scholar]
- Vernay B, Koch M, Vaccarino F, Briscoe J, Simeone A, Kageyama R, Ang SL (2005) Otx2 regulates subtype specification and neurogenesis in the midbrain. J Neurosci 25: 4856–4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY (2000) Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet 26: 229–232 [DOI] [PubMed] [Google Scholar]
- Xu W, Edmondson DG, Roth SY (1998) Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Mol Cell Biol 18: 5659–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Nagano T, Takehara S, Hibi M, Aizawa S (2005) Shisa promotes head formation through the inhibition of receptor protein maturation for the caudalizing factors, Wnt and FGF. Cell 120: 223–235 [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Yamauchi J, Kuwata T, Tamura T, Yamashita T, Bae N, Westphal H, Ozato K, Nakatani Y (2000) Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc Natl Acad Sci USA 97: 11303–11306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao TP, Oh SP, Fuchs M, Zhou ND, Ch'ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R (1998) Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93: 361–372 [DOI] [PubMed] [Google Scholar]
- Yao YL, Yang WM, Seto E (2001) Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol 21: 5979–5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 1 Legend
Supplementary Figure 2
Supplementary Figure 2 Legend
Supplementary Figure 3
Supplementary Figure 3 Legend
Supplementary Figure 4
Supplementary Figure 4 Legend
Supplementary Materials and Methods


