Abstract
Constitutive activation of the transmembrane receptor, Notch3, and loss of function of the hematopoietic transcription repressor, Ikaros (IK), play direct roles in T-cell differentiation and leukemogenesis that are dependent on pre-T-cell receptor (pre-TCR) signaling. We demonstrate the occurrence of crosstalk between Notch3 and IK that results in transcriptional regulation of the gene encoding the pTα chain of the pre-TCR. We also show that, in the presence of the pre-TCR, constitutive activation of Notch3 in thymocytes causes increased expression of dominantnegative non-DNA-binding IK isoforms, which are able to restrain the IK inhibition of Notch3's transcriptional activation of pTα. This effect appears to be mediated by Notch3's pre-TCR-dependent upregulation of the RNA-binding protein, HuD. Notch3 signaling thus appears to play a critical role in the diminished IK activity described in several lymphoid leukemias. By exerting transcription-activating and transcription-repressing effects on the pTα promoter, Notch3 and IK cooperate in the fine-tuning of pre-TCR expression and function, which has important implications for the regulation of thymocyte differentiation and proliferation.
Keywords: alternative splicing, Ikaros, Notch3, pre-TCR, T-cell leukemia
Introduction
The process of T-cell leukemogenesis is regulated by some of the same mechanisms that control T-cell differentiation, and the final outcome depends on fine balancing. Both processes are known to be affected by aberrantly activated Notch signaling and by the loss of function of the transcription factor, Ikaros (IK), depending on the presence of a functional pre-T-cell receptor (pre-TCR) (Winandy et al, 1995, 1999; Allman et al, 2001; Bellavia et al, 2002).
Our previous studies in transgenic mice expressing the intracellular domain (IC) of the Notch3 receptor (N3-IC) have shown that constitutive Notch3 signaling leads to the persistent overexpression in thymocytes of the pTα chain, an essential component of the pre-TCR. This overexpression is a hallmark of the T-cell leukemia/lymphoma that ultimately develops in these animals (Bellavia et al, 2000). Interestingly, by generating double mutant mice transgenic for Notch3-IC and lacking pTα (N3-IC/pTα−/− mice), we observed that an intact pre-TCR is required for T-cell leukemogenesis (Bellavia et al, 2002), although it is not required for Notch3-driven T-cell differentiation. Indeed, whereas pTα knockout arrests T-cell development, transgenic N3-IC overcomes this block, driving immature CD4−CD8− double-negative (DN) thymocytes to progress toward a more differentiated phenotype (e.g. acquisition of CD4, CD8 and TCRβ expression) despite the absence of a functional pre-TCR (Bellavia et al, 2002).
An intriguingly similar picture emerges when we analyze the effects of the loss of IK activity. Dominant mutation that eliminates the DNA-binding activity of this transcription repressor leads to rapid development of murine leukemia and lymphoma, which is strictly dependent on either pre-TCR or TCR signaling (Winandy et al, 1995), and yet IK-deficient DN thymocytes can differentiate to the CD4+CD8+ double-positive (DP) and CD4+ single-positive stages in the absence of a pre-TCR complex (Winandy et al, 1999). Therefore, like constitutive activation of Notch3, loss of IK function is characterized by pre-TCR-independent thymocyte differentiation from the DN to the DP stage and pre-TCR-dependent expansion of immature thymocyte populations (Winandy et al, 1999; Bellavia et al, 2002).
The roles of Notch and IK in human leukemogenesis are supported by several reports. Notch3 overexpression has been observed in virtually 100% of human T-cell acute lymphoblastic leukemia/lymphomas (ALL), including tumors from all major molecular and immunophenotypic subtypes (Bellavia et al, 2002), and activating mutations of Notch1 have been found in over 50% of these tumors (Weng et al, 2004). A high percentage of infant B- and T-ALLs also display an increased expression of short non-DNA-binding IK isoforms (Sun et al, 1999a, 1999b). Alternatively spliced transcripts of the ikaros gene encode at least nine protein isoforms (IK-1–9) with different DNA-binding capabilities (Hahm et al, 1994; Molnar and Georgopoulos, 1994; Beverly and Capobianco, 2003). Isoforms IK-1–3 are characterized by at least three N-terminal zinc-finger motifs that allow efficient DNA binding. Shorter isoforms, lacking one or more of these DNA-binding motifs, form heterodimers with full-length isoforms and exert dominant-negative effects that can decrease or even suppress normal IK activity. It has recently been suggested that increased expression of dominant-negative IK isoforms (IK-dn) and constitutively activated Notch play cooperative roles in leukemogenesis, involving effects that may converge in the transcriptional regulation of one or more key genes (Beverly and Capobianco, 2003). However, the identity of these putative common targets is still obscure, and thus far there has been no demonstration of a direct link between aberrant Notch signaling and alterated IK isoform expression.
In the present paper, we show that Notch3 activation upregulates the expression in thymocytes of the RNA-binding protein HuD. This effect results in the increased expression of IK-dn isoforms, which diminishes the DNA-binding activity of IK and, consequently, its ability to inhibit Notch3-induced increase of pTα expression. These events could be expected to lead to a persistent activation of pre-TCR signaling, which is responsible for the triggering of a number of oncogenic pathways.
Results
Notch3-IC transgenic mice display increased expression of alternatively spliced IK isoforms
To better define the relationships between IK and Notch3 in the processes of T-cell differentiation and leukemogenesis, we first analyzed IK isoform expression in premalignant thymocytes from young (2-week-old), wild-type (wt) and N3-ICtg mice. We define the line between preleukemia and leukemia by the finding of CD4+CD8+ DP cells in the spleen of transgenic mice. The two groups of cells were similar in terms of thymocyte subset distributions (Figure 1A), but the N3-ICtg cells displayed increased expression of IK-dn isoforms at both the RNA and protein levels (Figure 1B, upper and lower panels, respectively). We then investigated IK protein expression in adult N3-ICtg mice that had already developed unequivocal signs of leukemia/lymphoma (Bellavia et al, 2000). Thymocytes (Figure 2A, upper panels) and peripheral T-cells (i.e. lymph node cells) (Figure 2A, lower panels) from these animals exhibited significantly increased expression of the IK-dn isoforms (compared with wt littermates). The IK isoform profiles were unrelated to the immunophenotype of the lymphoma cell, which can vary from animal to animal (Bellavia et al, 2000). In fact, similar profiles were observed in cells with different immunophenotypes (e.g. N3-ICtg3 and N3-ICtg4 in Figure 2A and B), and those with identical phenotypes sometimes had different IK expression profiles (although the increase in IK-dn isoforms was a constant) (data not shown). It is interesting to note that in each tumor-bearing mouse, thymus- and lymph node-derived lymphoma cells consistently displayed identical immunophenotype (Figure 2A). Similar IK isoform profiles were also observed at protein levels (Figure 2B).
Figure 1.
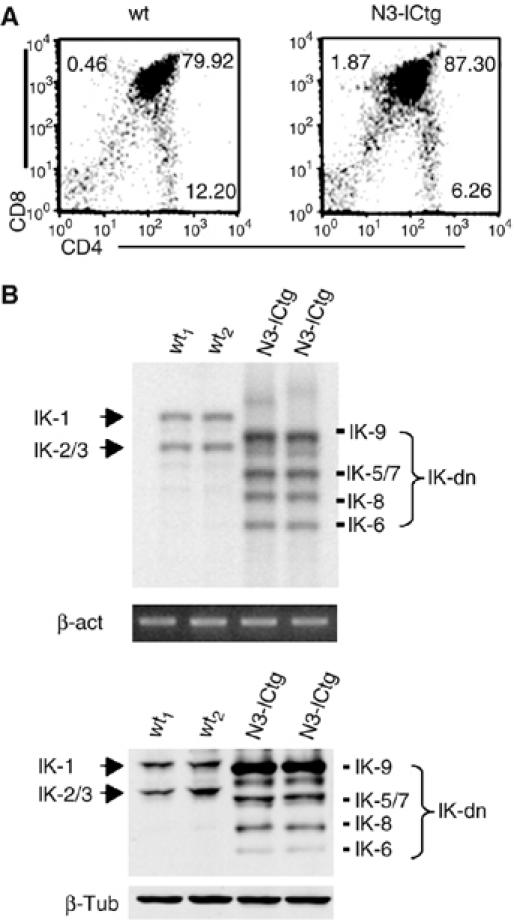
Alternatively spliced IK isoforms are overexpressed in premalignant thymocytes from Notch3-IC transgenic (N3-ICtg) mice. (A) CD4+ and/or CD8+ subset distributions (documented by two-color FCA) of thymocytes from 2-week-old wt and N3-ICtg mice. The number in each quadrant indicates the percentage of total cells represented by the corresponding subset. (B) IK isoform expression profiles assessed by RT–PCR (upper panel) and Western blot (lower panel) analysis of whole thymocyte lysates from two wt and two N3-ICtg mice. IK-1 and IK2/3: DNA-binding IK isoforms; IK-dn: dominant-negative IK spliced variants incapable of DNA binding. PCR products were analyzed by Southern blot using IK-6 cDNA as probe to detect the specific sequences. mRNA expression was monitored along the exponential phase of amplification and normalized to β-actin (β-act). In Western blot assay, results were normalized to β-tubulin (β-tub). The data are representative of three similar experiments.
Figure 2.
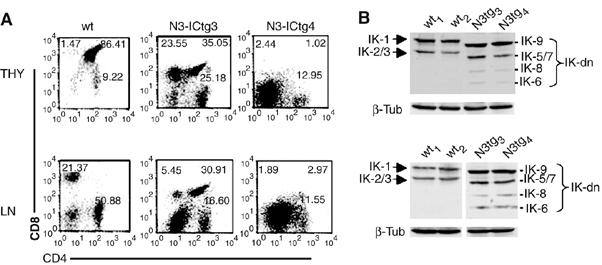
IK isoform expression patterns are altered in T lymphoma cells from Notch3-IC transgenic (N3-ICtg) mice. (A) Two-color FCA analysis of CD4 versus CD8 expression in freshly isolated thymocytes (THY) (upper panels) and lymph node cells (LN) (lower panels) from 5-week-old mice: two wild-type (wt1 and wt2) and two N3-ICtg (N3-ICtg3 and N3-ICtg4). The number in each quadrant indicates the percentage of total cells represented by the corresponding subset. (B) Immunoblots of whole-cell extracts from the cells shown in panel A. IK1 and IK2/3: DNA-binding IK isoforms; IK-dn: dominant-negative IK isoforms incapable of DNA binding.
Noteworthily, the IK-9 isoform, of intermediate size with respect to IK-1 and IK-2/3, appears mainly represented in all the samples obtained from N3-ICtg cells. This isoform, previously described by Beverly and Capobianco (2003), includes all the exons except exon 4, which encodes the critical DNA-binding zinc-fingers, and thus behaves as dominant negative. We cloned and sequenced IK isoforms from thymocytes of both Notch3-IC and wt mice, and Supplementary Figure 1 schematically represents the bands more represented, including the distribution of zinc-finger motifs and the size range of RNA.
The pTα/pre-TCR is required for Notch3-induced redistribution of IK isoforms
Previous work has shown that the presence of the pre-TCR is necessary for Notch3-induced leukemogenesis (Bellavia et al, 2002) and for that observed in mice with a dominant DNA-binding mutation in the IK gene (Winandy et al, 1995, 1999). We hypothesized that increased expression of IK-dn isoforms is characteristic of Notch3-dependent leukemia and may also contribute to its development. To investigate this possibility, we compared the IK isoform expression profiles of thymocytes from N3-ICtg mice, double mutant N3-ICtg/pTα−/− mice, pTα−/− mice and wt controls. DN (CD4−CD8−) and DP (CD4+CD8+) thymocytes were sorted and analyzed separately to minimize the effects of any strain-related differences in thymocyte subset distributions. As shown in Figure 3A, the shift toward IK-dn expression observed in N3-ICtg thymocytes was absent in both N3-ICtg/pTα−/− and pTα−/− cells, which lack a functional pre-TCR complex. Intriguingly, semiquantitative RT–PCR amplification of IK exon 7 (present in all of the isoforms) revealed that total IK transcript levels in DN and DP thymocytes from N3-ICtg/pTα−/− double mutant mice were significantly lower than those observed in wt, N3-ICtg and pTα−/− cells (Figure 3A, middle panels). These findings were confirmed at the protein expression level by Western blot analysis of total cell extracts from unfractionated thymocytes (Figure 3B).
Figure 3.
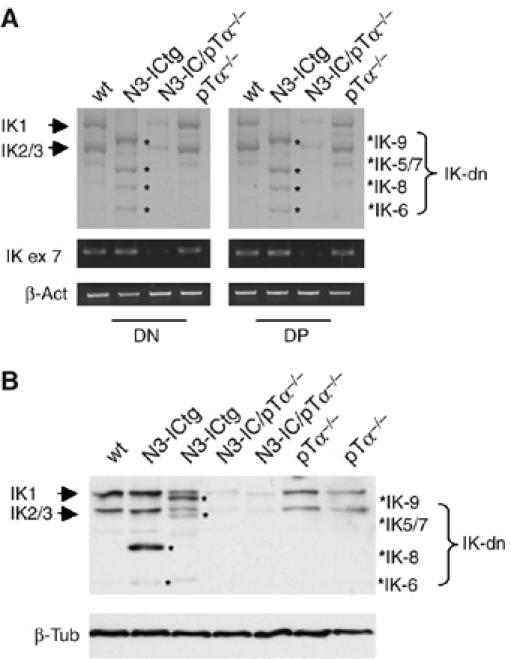
IK isoform expression patterns in thymocytes depend on pTa expression. (A) RT–PCR analysis of IK isoform expression was performed on RNA from sorted CD4-CD8− DN and CD4+CD8+ DP thymocytes from wild-type (wt), Notch3-IC transgenic (N3-ICtg), Notch3-IC/pTα−/− and pTα−/− mice. (B) IK immunoblot of whole-cell lysates of unfractionated thymocytes from one wt mouse and two mice of each of the three mutant genotypes listed above. IK1 and IK2/3: DNA-binding IK isoforms; IK-dn: dominant-negative IK isoforms incapable of DNA binding; IK ex 7: IK exon 7. Results were normalized to β-tubulin (β-tub).
Notch3 modulates expression patterns of IK mRNAs and proteins in vitro
Our next step was to determine whether Notch3 signaling directly affects IK isoform expression. The in vivo findings presented above indicated that such an effect is probably dependent on the presence of an intact pre-TCR. Based on CD25 and CD44 expression, DN thymocytes can be divided into four subsets, DN1–4, and functional pre-TCR expression is first observed at the DN3 (CD25+CD44−) stage. Therefore, we compared Notch3-induced IK splice variant profiles in DN1 and DN3 cells represented respectively by the M31 (Primi et al, 1988) and 2017 (Spolski et al, 1988) cell lines (Supplementary Figure 2). The two cell lines (both of which constitutively express full-length DNA-binding IK isoforms) were transiently transfected with increasing amounts of N3-IC, and IK isoform profiles were investigated 48 h later by semiquantitative RT–PCR and Western blotting. N3-IC-transfected 2017 cells displayed significantly increased expression of IK-dn isoforms (compared with untransfected or empty vector-transfected controls) at both the mRNA (Figure 4A, upper right panel) and protein (Figure 4B, right panel) levels, but there was no evidence of these isoforms in transfected M31 cells (Figure 4A, lower panels). The shift toward IK-dn expression seems to be a Notch3-specific effect as it was not observed after transient transfection of 2017 or M31 cells with Notch1-IC (Figure 4, left panels). Noteworthily, Notch1-IC was able to increase the expression of Hes-1 and Deltex in transfected 2017 cells (Figure 4A, upper left panels). Moreover, it was able to increase the Hes-1 promoter activity in a luciferase reporter assay to a greater extent than Notch3-IC in both HEK293 and M31 cells (Supplementary Figure 3). We finally addressed the possible effect of exogenous Notch1-IC on the expression of Notch3 in 2017 cells. The middle panel of Figure 4A shows that enforced expression of exogenous Notch1-IC does not modify the endogenous Notch3 expression of 2017 cells.
Figure 4.
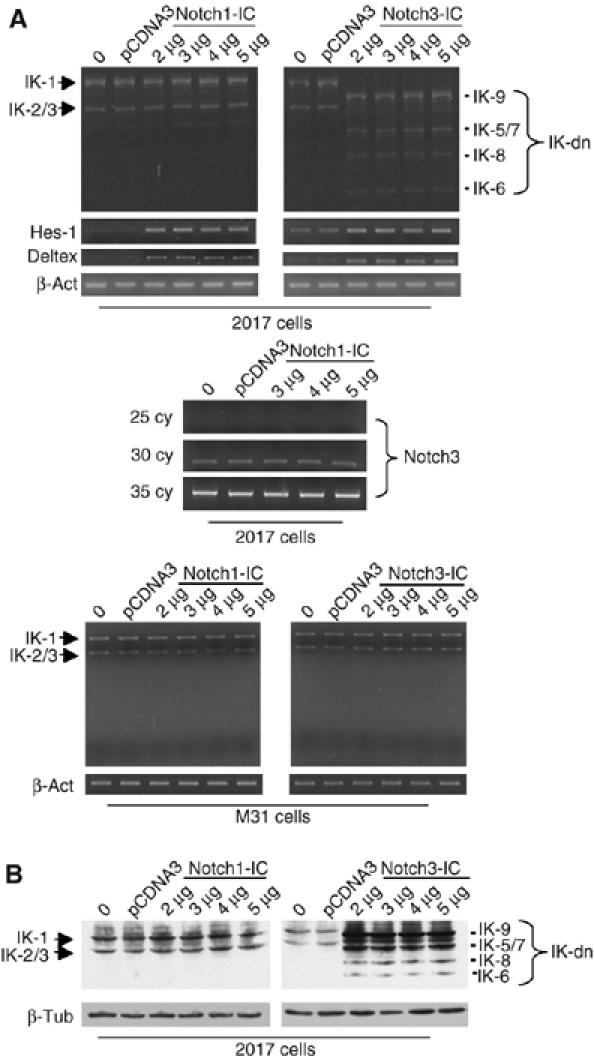
Notch3 regulates alternative splicing of IK (IK) in vitro. DN1 and DN3 thymocytes (represented respectively by the M31 and 2017 cell lines) were transfected with increasing amounts of Notch3 (right panels) or Notch1 (left panels), and IK mRNA and protein expression was assessed by RT–PCR (A) and Western blotting (B). In the upper panels (A), Hes-1 (30 cycles for Notch1 and 35 cycles for Notch3) and Deltex RNA expressions were used as control of correct function of Notch1-IC and Notch3-IC constructs. The middle panel (A) shows the inability of Notch1-IC to increase the constitutive expression of Notch3 by 2017 cells at any cycles considered. IK1 and IK2/3: DNA-binding IK isoforms; IK-dn: dominant-negative IK isoforms incapable of DNA binding. β-Actin (β-act) and β-tubulin (β-tub) are shown as controls of equivalent loading of each sample.
Collectively, the findings presented thus far indicate that activated Notch3 signaling in the presence of pre-TCR expression can modulate IK isoform expression profiles in vivo and in vitro. This effect, which appears to be Notch3-specific, could thus be responsible (at least in part) for the abnormal expression of IK-dn isoforms in premalignant and malignant thymocytes of N3-ICtg mice.
Notch3 and IK act cooperatively to regulate pTα gene expression
The findings described above, together with a recent report that loss of IK function leads to increased expression of Notch3 and pTα in murine T lymphoma cells (Dumortier et al, 2006), suggest the possibility of direct crosstalk between Notch3 and IK that serves to regulate pTα gene transcription. Previous studies have shown that the pTα enhancer is activated by Notch1 signaling via CSL-binding sites (Reizis and Leder, 2002), and we have demonstrated dose-dependent activation of the pTα promoter by Notch3 (Talora et al, 2003). To further elucidate the mechanisms underlying the effects of Notch3 on the regulatory region of pTα, we transfected non-lymphoid HEK293 cells with a luciferase reporter construct containing the entire pTα promoter sequence (as reported in GenBank, accession number U27268). Enforced coexpression in these cells of N3-IC and Mastermind (MAM), the positive regulator of Notch activity, increased pTα promoter activity ∼10-fold, and additional increases were observed after the addition of RBP-Jκ/CSL (Figure 5 and not shown). Although the ability of Notch-IC to activate transcription is known to be potentiated by its interaction with these coactivators (Wu et al, 2000; Fryer et al, 2002; Jeffries et al, 2002; Lin et al, 2002), this is the first demonstration that Notch3-IC-driven activation of the pTα regulatory element is significantly enhanced by a transcriptional complex composed of RBP-Jκ/CSL and MAM.
Figure 5.
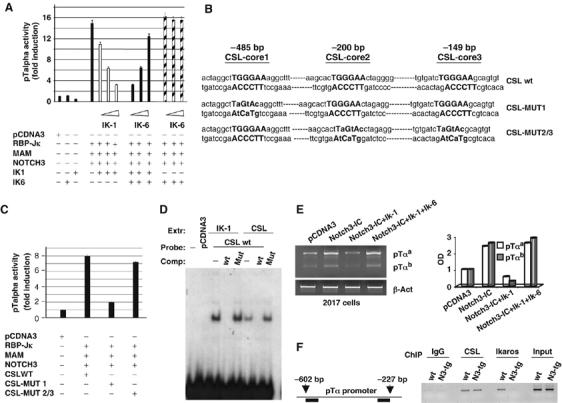
Transcriptional activation of the pTα promoter by Notch3 is differentially regulated by IK isoforms. (A) HEK 293 cells were transfected with a luciferase reporter construct containing the pTα promoter together with Notch3, RBP-Jκ, and Mastermind (MAM). The pTα promoter is activated by the Notch3 transcription activator complex (Notch3+RBP-Jκ+MAM). Increasing amounts of IK-1 alone (200–800 ng) dose dependently repressed Notch3-driven activation of the pTα promoter, but the non-DNA-binding IK6-dn isoform alone (200–800 ng) had no impact. When IK-1 and IK-6 were cotransfected, increasing amounts of IK-6 progressively diminished the transcription repression induced by fixed amount of IK-1. DNA contents in the different transfection assays were normalized with empty vector (pcDNA3). The data shown were collected from three independent experiments; vertical bars indicate standard deviation. (B) Mutational analysis of the TGGGAA sequence in the pTα promoter. Wild-type and mutated sequences from the entire promoter are shown in boldface type and the cloned CSL wt sequence and mutant constructs, CSL-MUT1 and CSL-MUT2/3, are shown. (C) Luciferase activity of wild-type and mutant pTα promoter constructs. pCDNA3: empty vector. The histograms represent mean results of three independent transfection experiments. Vertical bars indicate the range of standard errors. (D) EMSA: nuclear extracts derived from COS cells transfected with either IK-1 (IK1) or CSL (CSL) expression vectors were probed with an oligo containing the wild-type CSL-core1 consensus (TGGGAA) (CSL-wt), as indicated in (B). For competition (comp), 100-fold molar excess of cold CSL-wt or CSL-MUT 1 was used. (E) RT–PCR expression analysis of pTαa and pTαb isoform mRNAs in 2017 cells. The cells were transiently transfected with Notch3-IC alone to induce increased pTα expression or together with Ik-1 or Ik-1+Ik-6 to demonstrate that Notch3-dependent induction is repressed by Ik-1 and derepressed by Ik-6. pTα mRNA levels were normalized to β-actin (β-act) (left panel) and optical densities relatively to empty vector-transfected cells (pCDNA3) are shown (right panel). (F) Schematic representation of pTα promoter region, assessed by ChIP is indicated (left panel). Primary thymocytes derived from wt and Notch3-IC transgenic mice (N3-tg) were processed for chromatin preparation and then subjected to immunoprecipitation with antibodies against RBP-Jk (CSL), IK or IgG as control. Immunoprecipitated DNA was analyzed by PCR with pTα promoter specific primers (right panel). The results are representative of three similar experiments.
Full-length DNA-binding IK isoforms have been shown to repress CSL-dependent Notch-triggered transcriptional activity, and this effect is attenuated by the coexpression of alternatively spliced IK-dn isoforms (Beverly and Capobianco, 2003). Similarly, enforced expression of the IK-1 isoform in HEK 293 cells significantly inhibited activation of the pTα promoter triggered by the Notch3 and coactivator complex (Figure 5A), but there was no evidence of transcriptional repression in cells transfected with the dominant-negative IK-6 alone (Figure 5A). In the presence of a constant level of coexpressed IK-1, the increasing presence of IK-6 progressively diminished and ultimately overcame the transcriptional repression induced by the full-length isoform, restoring pTα promoter activation to levels similar to those produced by the Notch3-dependent transcriptional complex in the absence of IK (Figure 5A). This body of evidence strongly suggests that Notch3 regulates the alternative splicing of IK and that, in so doing, it modulates its own ability to induce transcriptional activation of pTα.
It remained to be seen how the Notch3-triggered complex interacts with the pTα promoter region and what is the basis for IK antagonism. Our analysis revealed that the pTα promoter contains a canonical consensus CSL-binding sequence, several consensus sequences that are specific for IK and three TGGGAA motifs corresponding to the RBP-Jk/CSL core sequence. The latter finding was particularly interesting as this same core sequence is also present in the consensus DNA-binding sequences of IK (Molnar and Georgopoulos, 1994; Beverly and Capobianco, 2003). The three motifs were designated CSL-core 1, CSL-core 2 and CSL-core 3 in accordance with their positions in the pTα promoter (−485, −200 and –149 bp, respectively) (Figure 5B). The pTα promoter region containing the three motifs was cloned and to evaluate their possible roles in Notch3-dependent transcriptional activation, we generated two luciferase reporter constructs, one (CSL-Mut1) with a specific mutation in CSL-core1 and the second (CSL-Mut2/3) with mutations in CSL-core2 and CSL-core3 (Figure 5B), and transfected them, together with N3-IC and its coactivators, into HEK293 cells. In cells expressing the CSL-Mut2/3, luciferase activity was similar to that observed with the wt sequence, but CSL-Mut1 expression was associated with drastically reduced activity (Figure 5C). These findings indicate that the TGGGAA sequence at −485 bp is necessary for Notch3-dependent transcriptional activation of pTα.
In order to study the basis for Notch3 and IK-1 antagonism on pTα transcriptional activation, we first examined the ability of both IK-1 and CSL to bind the same site of pTα regulatory sequence by using the CSL wt sequence of the pTα promoter in an electrophoretic mobility shift assay (EMSA). We observed that both the IK-1 and CSL proteins, obtained from nuclear extracts derived from COS cells transfected with either IK-1 or CSL expression vectors, were able to bind the same site. We used as probe an oligo containing the wt CSL-core1 consensus (TGGGAA) of the pTα promoter, as indicated in Figure 5B. The interactions of IK-1 and CSL with the oligo were specific, as they were able to be competed with 100-fold molar excess of the unlabeled CSL-wt, but not with 100-fold molar excess of unlabeled mutant oligo CSL-MUT 1 (Figure 5D).
Moreover, the luciferase reporter and EMSA assays were confirmed in Notch3-IC transiently transfected 2017 cells by evaluating the ability of Notch3-IC to induce an increase in endogenous pTα expression and demonstrating that this induction is repressed by IK-1 and derepressed by IK-6 (Figure 5E).
Finally, to address whether IK and CSL truly compete in vivo for the same candidate site in the pTα promoter, we performed chromatin immunoprecipitation (ChIP) experiments (Saccani and Natoli, 2002) using primary thymocytes derived from wt and N3-ICtg mice. Figure 5F (right panel) shows that both CSL and IK proteins are associated with the pTα promoter in chromatin extracts derived from wt thymocytes. In contrast, CSL but not IK antibodies efficiently recovered the pTα promoter fragment using chromatin extracts derived from N3-ICtg mice. These results demonstrate that CSL and IK directly associate at the same binding site in the pTα promoter and might explain the functional collaboration observed between Notch3 and the IK-dn isoforms in sustaining pTα expression, finally supporting the hypothesis that the increased amount of IK-dn isoforms in thymocytes of N3-ICtg mice prevents the DNA binding of IK, thus allowing the derepression of pTα expression.
The effects of Notch3 on the expression of IK spliced variants depend on its pre-TCR-dependent upregulation of the RNA-binding protein HuD
Next, we attempted to identify mechanisms specifically triggered by Notch3 that would account for the presence of alternatively spliced IK isoforms. In a recent Affymetrix MAS 5.0 microarray analysis of the global gene expression profile of N3-ICtg premalignant thymocytes (results currently unpublished), we discovered a striking increase (∼39-fold compared with wt controls from 2-week-old mice) of mRNA for HuD, an RNA-binding protein of the ELAV/Hu family (Inman et al, 1998), which is reportedly capable of regulating both the stability (Levine et al, 1993; Mobarak et al, 2000; Deschenes-Furry et al, 2003; Ratti et al, 2006) and the alternative splicing (Lisbin et al, 2001; Zhu et al, 2006) of RNA. In the present study, we confirmed, by means of semiquantitative RT–PCR the presence of significantly increased HuD mRNA levels in DN and DP thymocytes from N3-ICtg mice (Figure 6A). Even more interesting was our finding that Notch3-induced enhancement of HuD expression is pre-TCR dependent. In fact, in contrast to wt and N3-ICtg thymocytes, cells from both Notch3-IC/pTα−/− and pTα−/− mice displayed HuD expression levels that were virtually undetectable in both DN and DP cells (Figure 6A). In addition, HuD expression was dose-dependently upregulated in the pre-T cell line 2017 after transient transfection with increasing amounts of Notch3-IC (Figure 6B, left panel), but no increases were observed in transfected M31 cells (not shown). The negative results observed when transfections were carried out with Notch1-IC (Figure 6B, right panel) indicate that the upregulation of HuD is a Notch3-specific effect.
Figure 6.
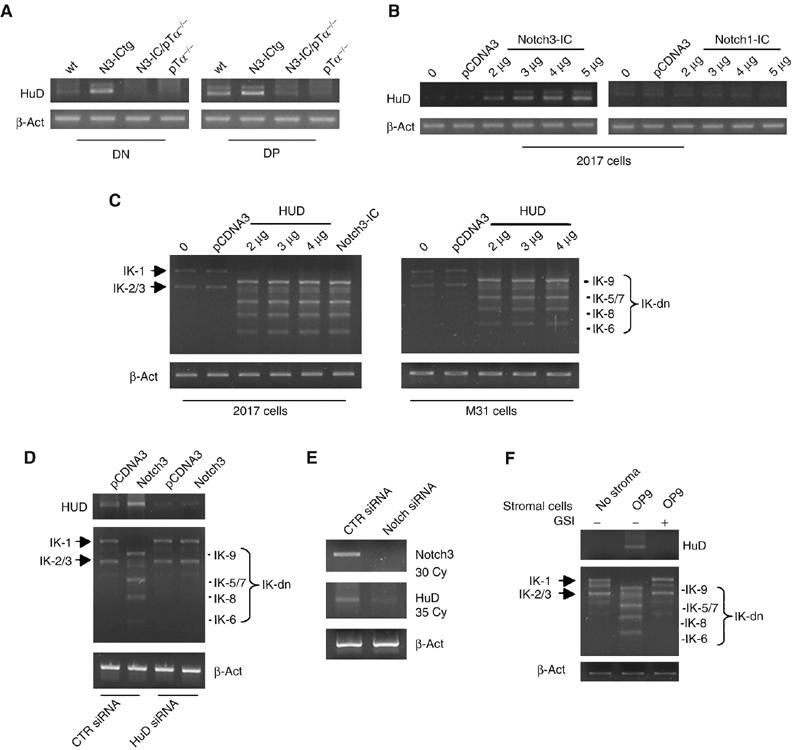
Notch3-induced skewing of Ikaros (IK) isoform profiles is mediated by Notch3-dependent upregulation of the RNA-binding protein, HuD. HuD expression was assessed in vivo by semiquantitative RT–PCR analysis of RNA in (A) sorted CD4−CD8− DN and CD4+CD8+ DP thymocytes from wt, N3-ICtg, N3-ICtg /pTα−/− and pTα−/− mice and (B) in vitro in 2017 cells transfected with increasing amounts of Notch3 (left panel) or Notch1 (right panel). (C) RT–PCR detected expression of alternatively spliced IK isoforms in 2017 and M31 cells transfected directly with HuD. (0=non-transfected cells; pCDNA3=empty vector-transfected cells.) (D) HuD siRNA assay showing that knockdown of endogenous HuD prevents the Notch3-induced modulation of IK-dn isoforms. HuD and IK mRNA expression profile as assessed by RT–PCR in 2017 cell line, transfected with pCDNA3 or Notch3 expression vectors, after HuD or scrambled (CTR) siRNA is shown. (E) siRNA of Notch3 is able to inhibit the constitutive expression of HuD in 2017 cells. RT–PCR for HuD and Notch3 mRNAs in 2017 cell line, after HuD or scrambled (CTR) siRNA is shown. (F) Coculture experiment of 2017 cells on OP9 stromal cells. RT–PCR for HuD (30 cycles) and IK mRNAs is shown. GSI: γ-secretase inhibitor I; IK-1 and IK-2/3: DNA-binding IK isoforms; IK-dn: dominant-negative IK isoforms incapable of DNA binding. mRNA expression was monitored along the exponential phase of amplification and normalized to β-actin (β-act).
Collectively, these results strongly suggest that the Notch3-induced upregulation of HuD expression, which is pre-TCR-dependent, may be related to the increased generation and/or enhanced stability of IK-dn isoforms. As a putative downstream effector of pre-TCR signaling, HuD expression alone would be expected to allow the shift toward IK-dn isoform expression even in the absence of pre-TCR. This hypothesis was confirmed when we transiently transfected 2017 and M31 cells with a pcDNA3 expression vector containing cloned HuD. In both cell lines, increasing amounts of transfected HuD elicited progressive increases in short IK-dn isoforms, an effect that was unrelated to the differentiation stage or to pre-TCR expression (Figure 6C).
Together, the above results suggest that HuD protein mediates the effect of Notch3 on the differential expression of IK isoforms. In order to directly address this issue, we performed siRNA experiment to study the effect of HuD silencing on the Notch3-induced modulation of IK-dn isoforms. Figure 6D shows that the silencing of HuD abrogates the effect of Notch3 on IK isoform modulation. We also show the reverse by silencing endogenous Notch3. Indeed, Figure 6E shows that the siRNA of Notch3 is able to inhibit the constitutive expression of HuD in 2017 cells.
It remained to be seen whether the HuD-increased expression and the shift toward IK-dn isoforms were induced by endogenous Notch signaling. As shown in Supplementary Figure 2, 2017 cells constitutively express endogenous pTα, Notch3 and Notch1, whereas M31 cells express similar levels of Notch1, but very low levels of Notch3 and undetectable pTα when compared with 2017. Thus, we performed coculture experiments of 2017 cells on OP9 stromal cells that have been described to constitutively express the Notch ligand Jagged-1 at levels similar to those of normal thymic epithelial cells and to be able to trigger endogenous Notch signaling (Schmitt and Zúñiga-Pflücker, 2002; Lehar et al, 2005). We observed that the coculture with OP9 cells induced the same effect on HuD and IK-dn isoform expression as the transfection of Notch3-IC, and when the γ-secretase inhibitor I was added to the culture medium the effect was inhibited, thus demonstrating that it was due to the triggering of endogenous Notch signaling (Figure 6F).
The RNA-binding protein HuD is expressed in human T-ALL
Whereas the expression of alternatively spliced IK isoforms has been previously reported in several human leukemias (Sun et al, 1999a, 1999b and 1999c; Olivero et al, 2000), HuD expression was not addressed before. The observations reported above prompted us to analyze HuD mRNA expression in human T-ALL, as well as the possible role of HuD and Notch3 in regulating the IK isoform expression pattern. We utilized the previously described human T-ALL cell line Molt-3 (Sun et al, 1999a). Figure 7A shows that this cell line, similar to the mouse 2017 cell line, constitutively expresses Notch1 and 3, HuD, both the pTα isoforms a and b and an altered IK isoform profile. In order to address directly the relationships between Notch, HuD and IK isoforms in human T-ALL, we performed siRNA experiments in Molt-3 cells. Figure 7B shows that knockdown of Notch3 and HuD but not of Notch1 is able to alter the IK isoform profile displayed by Molt-3 cells. Interestingly, besides specifically affecting the pattern of IK isoform expression, the knockdown of Notch3 results in a higher inhibition of proliferation of Molt-3 cells when compared with the knockdown of Notch1 (49 versus 30%) (Figure 7C).
Figure 7.
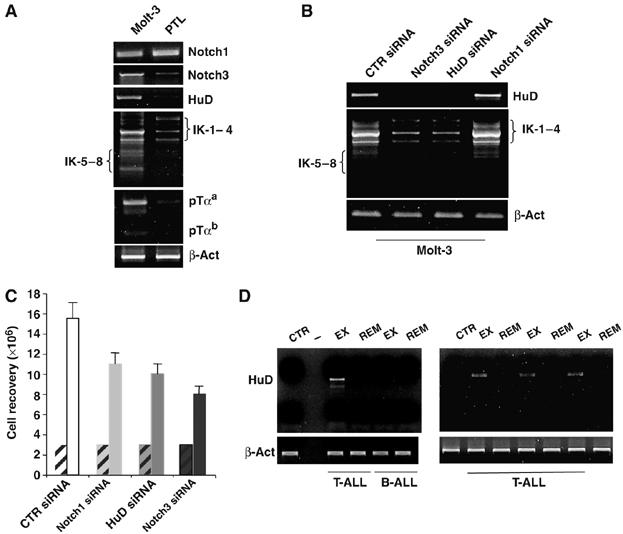
The RNA-binding protein HuD is expressed in human T-ALL and regulates IK isoform profile. (A) RT–PCR expression analysis of human Notch1, Notch3, HuD, IK and pTα mRNAs in Molt-3 cell line and peripheral T lymphocytes from one healthy donor (PTL) used as control. Results were normalized to β-actin (β-act). (B) siRNA assay showing that HuD and Notch3, but not Notch1, are able to regulate IK mRNA expression profile in Molt-3 cells, as assessed by RT–PCR; CTR siRNA: scrambled siRNA. (C) Inhibition of proliferation of Molt-3 cells. Recovery of Molt-3 cells after Notch3, HuD, Notch1 or scrambled siRNA transfection, compared to the cell number at the starting point (hatched bars). The data shown represent the average of three independent experiments; vertical bars indicate standard deviation. (D) HuD expression was assessed by semiquantitative RT–PCR analysis of RNA in bone marrow samples from four different primary human T-ALLs and one primary B-ALL at different stages of disease (exordium, EX; and remission, REM) and one control patient (CTR). Results were normalized to β-actin (β-act).
Finally, we studied four different primary human T-ALLs and one primary B-ALL utilizing bone marrow samples from patients at different stages of disease (exordium and remission) and one control patient. We observed expression of HuD in all of the T-ALL exordium samples; in contrast, HuD expression was undetectable in all the samples of the same patients in remission stage, in the sample from the B-ALL-bearing patient and in the control (Figure 7D).
Discussion
Pre-TCR expression is indispensable for the early steps of T-cell differentiation and for progression of Notch3-dependent T-cell leukemia (Fehling et al, 1997; Bellavia et al, 2002). The pTα gene encodes an essential pre-TCR component and is a key target of Notch3 signaling, which strongly enhances pTα promoter transcription through the formation of an activator complex composed of N3-IC, CSL and MAM. Our findings argue that the hematopoietic cell-specific transcription repressor, IK, is an important regulator of Notch3's effects on pTα. The pTα promoter region contains three consensus DNA-binding TGGGAA sequences that are recognized by both IK and the Notch effector protein CSL, and one of these was shown to be essential for Notch3-induced activation of pTα. This activation is inhibited by DNA-binding IK isoforms, but our findings indicate that, in the presence of a functional pre-TCR complex, Notch3 can eliminate this restraint by increasing the expression of IK-dn splice variants that are incapable of DNA binding. It appears, therefore, that, in addition to the direct effect exerted by its activator complex, Notch3 signaling also enhances pTα transcription indirectly by diminishing the transcription-repressing activity of IK.
The effect of Notch3 on IK isoform expression seems to be mediated, at least in part, by the RNA-binding protein, HuD. The highly conserved Hu proteins (and HuD in particular) play critical roles in the post-transcriptional modulation of gene expression, ranging from regulation of alternative splicing and translation to modulation of mRNA transport and stability (Levine et al, 1993; Mobarak et al, 2000; Lisbin et al, 2001; Deschenes-Furry et al, 2003; Ratti et al, 2006; Zhu et al, 2006). HuD has long been known as one of the earliest markers of neural differentiation (Perrone-Bizzozero and Bolognani, 2002), and more recently it has also been described in the CD34+ subset of bone marrow cells (Goolsby et al, 2003), but our findings are the first evidence of its expression by mouse thymocytes and, more importantly, in human T-ALL. Whereas low levels were observed in wt DP thymocytes, DN and DP thymocytes from N3-ICtg mice display high levels of HuD expression. Moreover, N3-IC transfection resulted in significantly increased dose-dependent expression of HuD. Even more intriguing was the correlation observed between this upregulation of HuD (which was strictly pre-TCR-dependent in vivo and in vitro) and the shift toward increased generation of IK-dn isoforms in cells. The fact that the same shift was also observed following transfection of HuD alone, independently of pre-TCR presence, being instead inhibited by siRNA-induced silencing of endogenous HuD, adds further support to the view that this protein plays a direct role in Notch3's modulation of IK isoform expression patterns. Moreover, the decreased expression of HuD following the knockdown of endogenous Notch3 by siRNA and its increased expression observed after coculturing 2017 cells on Jagged-1-expressing OP9 cells, inhibited by the treatment with GSI, suggest that HuD expression is induced by and depends on the triggering of endogenous Notch signaling.
The virtual absence of IK-dn isoforms in N3-IC/pTα−/− double mutant thymocytes was accompanied by a decrease in IK-1 and IK-2/3 transcripts as well (with respect to levels found in N3-ICtg, pTα−/− and wt cells). This finding suggests that the absence of HuD might also affect IK expression at a pre-mRNA level. More intriguingly, the observation that HuD is not expressed in the absence of pTα, independently of the overexpression of Notch3, suggests that Notch signaling alone, in the absence of pTα, is not sufficient in sustaining HuD expression.
Reduced expression of IK-1 and 2/3, by decreasing the inhibitory role of IK DNA-binding isoforms, could conceivably potentiate Notch3 activity and favor the activation of Notch target genes other than pTα, and an effect of this type might explain Notch3's partial reversal of the developmental arrest that characterizes thymocytes from pTα−/− single mutant mice (Bellavia et al, 2002). IK target/binding sites have also been reported in the enhancer region of CD3 and CD4 genes (Molnar and Georgopoulos, 1994).
In conclusion, we provide evidence for a novel non-redundant mechanism whereby the transmembrane receptor Notch3 and the hematopoietic nuclear factor IK cooperate in the regulation of T-cell development and lymphomagenesis. The molecular model portrayed by our findings (Figure 8) is characterized by crosstalk among Notch3, IK and the pTα/pre-TCR signaling activity that is mediated in part by the RNA-binding protein HuD. Its pre-TCR-dependent upregulation, specifically induced by Notch3, diminishes the IK-induced repression of transcriptional activation of pTα, an effect that appears to be related to regulation of the differential splicing and/or the stability of IK mRNAs. This is the first evidence that the RNA-binding protein HuD plays a role in the complex scenarios of T-cell differentiation and tumorigenesis.
Figure 8.
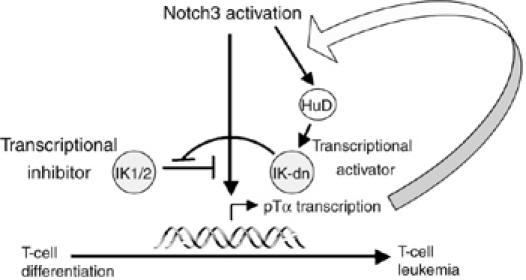
Crosstalk among Notch3, IK and pre-TCR in the regulation of T cell development and lymphomagenesis. Overexpression of HuD by thymocytes is triggered in a pre-TCR-dependent manner by the activation of Notch3 signaling. As a result of its modulating effects on the splicing and/or stability of IK mRNA, HuD promotes the preferential expression of IK-dn isoforms, which diminish IK-induced transcriptional repression and further enhance the upregulation of pTα gene expression induced directly by the Notch3–CSL transcription activator complex.
Materials and methods
Mice
The generation and typing of N3-ICtg (Bellavia et al, 2000), pTα−/− (Fehling et al, 1995) and N3-IC/pTα−/− double mutant mice (Bellavia et al, 2002) have been described elsewhere.
Flow cytometry analysis
Freshly isolated cells from thymi and lymph nodes were prepared and stained as previously described (Bellavia et al, 2000) and analyzed on a FACScan (BD Biosciences, Mountain View, CA) using CellQuest software (BD Biosciences). Forward and side scatter gatings were used to exclude dead cells from the analysis. Cells were stained with anti-CD4-FITC and anti-CD8-PE antibodies (BD PharMingen). PE- and FITC-conjugated rat IgG (BD PharMingen) were used as a control for immunofluorescence.
Cell sorting
Thymocyte suspensions from wt, Notch3-IC tg, Notch3-IC/pTα−/− and pTα−/− mice were prepared and stained with anti-CD4-FITC and anti-CD8-PE antibodies as described above. CD4−CD8− DN and CD4+CD8+ DP subsets were then separated (purity level ⩾95%) with the FACSAria cell sorter (BD Biosciences).
RT–PCR analysis
Total RNA was extracted from unfractioned T lymphocytes (both from thymi and lymph nodes), from CD4−CD8− and CD4+CD8+ sorted thymocytes, from untransfected and transfected M31, pre-T-cell line 2017 (Spolski et al, 1988) and human Molt-3 cell line, and from different samples of human ALLs and relative controls, using Trizol (Gibco) following the manufacturer's protocol. A 1 μg portion of RNA was processed for RT–PCR as previously described (Felli et al, 1999). PCR was performed at the appropriate annealing temperature with the following primers: IK 5′-CTCCAGATGAAGGG GATGAG-3′ and 5′-CAGCAGCAGCAAGTTATCCA-3′, IK exon 7 5′-TGGTTATCACAGCCAGGACA-3′ and 5′- AAATCAAACGCCAAAC AACC-3′; HuD 5′-AGAAGGGAATGTCAGCTTTT-3′ and 5′-TGAA TTCCTCTTGGGTCATA-3′ (Akamatsu et al, 2005); β-actin 5′-GTGG GCCGCTCTAGGCACCAAT-3′ and 5′- CTCTTTGATGTCACGCACGA TTTC-3′. Human IK: 5′-CCCCCTGTAAGCGATACTCCAGAT-3′ and 5′-GGCTTGGTCCATCACGTGGGGA-3′; human HUD: 5′-GAGTCTCTT CGGGAGCATTG-3′ and 5′-CTTGTGGGCTTTGTTGG TTT-3′; mouse Hes-1: 5′-ATGCCAGCTGATATAATGGAG-3′ and 5′-CACGCTCGGG TCTGTGCTGAG-3′; mouse Deltex: 5′-CACTGGCCCTGTCCACCCAG CCTTGGCAG-3′ and 5′-ATGCGAATT CGGGAAGGCGGGCAACTCAG-3′; mouse Notch3: 5′-CCAGGGCTGC AACACTGAGGAATG-3′ and 5′-TTGTGGCCAGCAGCTATGTCCTTG-3′; mouse pTalpha: 5′-GCGAT GCTTCTCCACGAGTGGGC-3′ and 5′-GCGCTATGTCCAAATTCTGT GGG-3′; human β-actin: 5′-CTACAATG AGCTGCGTGTGG-3′ and 5′-CGGTGAGGATCTTCATGAGG; mouse Notch1 : 5′-GTGGATGCTGACT GCATGGATGTC-3′ and 5′-ATGCAAAGCCGACTTGCCTAGGTC-3′; human Notch1: 5′-CTACCTGTCA GACGTGGCCT-3′ and 5-CGCAGA GGGTTGTATTGGTT-3′; human Notch3: 5′-TTCTTAGATCTTGGGGG CCT-3′ and 5′-GGAAGAAGG AGGTCCCAGAC-3′; human pTα: 5′-CTGCAGCTGGGTCCTGCCTC-3′ and 5′-AGTCTCCGTGGCCGGGTG CA-3′. To quantify transcript expression levels, PCR was carried out in the linear exponential phase of amplification through 20–35 cycles. Sample loading was monitored by a β-actin transcript that was subjected to the same treatment. IK PCR products were analyzed by agarose gel electrophoresis followed by Southern blotting and hybridization with a probe specific to the amplified sequences.
Western blot analysis
Whole-cell extracts were prepared as described previously (Beverly and Capobianco, 2003). In brief, 50 μg of protein extraction from each sample was subjected to SDS/PAGE, transferred to nitrocellulose membrane and probed with the following antibodies: anti-IK (SC-9861, Santa Cruz Biotechnology Inc.) and anti-tubulin (SC-8035, Santa Cruz Biotechnology Inc). Bound antibodies were detected with enhanced chemiluminescence (ECL kit, Amersham).
Cell lines
Transient transfection experiments were performed using the pre-T-cell line 2017 (Spolski et al, 1988) and M31 immature T-cell line (Primi et al, 1988).
Plasmids
IKcDNAs were cloned from RT–PCR reactions from both wt and Notch3-IC transgenic thymi as previously suggested (Beverly and Capobianco, 2003). The fragment containing the putative pTα promoter region (pubMed sequence U27268) was amplified by PCR from genomic DNA using the primers pTα-prom-FL 5′-GCTTTGGATCTGGAGGATGA-3′ and 5′-CTCTGCTAGTCTGCCTCC CA-3′ and subcloned in topo TA-cloning (Invitrogen, Carlsbad, CA, USA). The pTα promoter-luciferase fusion plasmids were constructed by cleaving with XhoI and SacI the PCR products and inserted into the XhoI and SacI sites upstream of the luciferase cDNA of pGL3-basic vector (Promega, Madison, WI, USA). The mutant constructs pTα-prom-FL CSL1MUT and pTα-prom-FL CSL2MUT+CSL3MUT were prepared using QuikChangeTM site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) according to the instructions of the manufacturer. MAM was cloned in pFLAG-CMV2 as previously suggested (Wu et al, 2000; Beverly and Capobianco, 2003). HuD was amplified by PCR by using the primers HuD 5′-GGGTCCATCTTCTGATCACA-3′ and 5′-GGGTGAGAAATT CAGGATTT-3′ and cloned in pcDNA3.1/V5-His-TOPO (Invitrogen, Carlsbad, CA, USA) expression vector. All the correct clones were confirmed by sequencing. The expression vectors for Notch3-IC (Bellavia et al, 2000) and RBP-Jκ (Talora et al, 2002), were previously described.
Electrophoretic mobility shift assay
IK-1 and CSL proteins were produced in transfected Cos cells using expression vectors containing IK-1 or CSL cDNA. Nuclear extracts were prepared (according to Dumortier et al, 2006) by resuspending 107 cells in 500 μl of lysis buffer (10 mM HEPES, pH 7.9; 1.5 mM MgCl2; 10 mM KCl; and 0.5 mM dithiothreitol (DTT)). After 10 min, cells were vortexed and nuclei were pelleted and resuspended in 50 μl of the following buffer: 20 mM HEPES, pH 7.9; 25% glycerol; 420 mM NaCl; 1.5 mM MgCl2; 0.2 mM EDTA; 0.5 mM DTT; and protease inhibitor cocktail. Lysates were kept on ice for 20 min and vortexed thoroughly. After centrifugation, supernatants (nuclear extracts) were quantified using the Bradford colorimetric assay. Three micrograms of nuclear extract was used for each sample. Samples were incubated for 25 min at room temperature with 2 μg of poly(dI-dC), 1 μg of bovine serum albumin and 10 μM ZnCl2 in 19 μl of HGDE buffer (20 mM HEPES, pH 7.9; 0.2 mM EDTA; 20% glycerol; 100 mM KCl; and 1 mM DTT). End-labeled, double-stranded probe was then added to each reaction mixture and the mixture was incubated for 20 min at room temperature. Protein–DNA complexes were resolved on a 5% polyacrylamide gel, dried and analyzed by autoradiography. The following probes were used: CSL: 5′-AGGAACTAGGCTTGGGAAAGGCTTTGAGAAT-3′; CSL MUT1: 5′-AGGAACTAGGCTTAGTACAGGCTTTGAGAAT-3′.
siRNA silencing
Mouse and human Notch3, HuD and control and human Notch1 siRNA (Dharmacon, Lafayette, CO, USA) were incubated in OPTI-MEM (Gibco, Gaithersburg, MD, USA) for 10 min at room temperature with Hy-perfect transfection reagent (Qiagen, Hilden, GE) at a final siRNA concentration of 5 nM. Complexes thus formed were added dropwise onto the cells (4 × 105 per well in a 24-well plate in 0.5 ml of RPMI medium containing FBS and antibiotics) and incubated for 72 h after transfection. To study Molt-3 cell survival/proliferation after siRNA, 3 × 106 cells/well were plated in a six-well plate; 72 h later, total cell recovery was evaluated.
Coculture experiments
OP9 stromal cells, kindly provided by Dr A Rolink, were mantained in ISCOVE'S modified supplemented medium, supplemented with 20% fetal bovine serum, 2% penicillin, 1% L-glutamine and 2% sodium pyruvate, and plated 1 day before use to achieve a confluent monolayer of cells. Cocultures were initiated with 4 × 106 2017 cells with or without 10 μM of γ-secretase inhibitor I (alternate name Z-LLNLe-CHO; Calbiochem, San Diego, CA, USA) and harvested after 48 h for RNA extraction.
Cell transfection and luciferase assays
Trasfection of the pre-T-cell line 2017 and M31 cell line was performed using the lipofectamine 2000 kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Renilla luciferase reporter vector pTK-Renilla-Luc (0.5 ng) was also incorporated into each transfection (Promega, Madison, WI, USA) for normalization. The total amount of transfected DNA was kept constant by adding empty vector. At 48 h post-transfection, the cells were lysed in a reporter lysis buffer (Passive Lysis Buffer; Promega, Madison, WI, USA) at 120 μl/well. Firefly- and pRL-TK-derived Renilla luciferase activities were measured in each sample with the Dual luciferase Assay System (Promega, Madison, WI, USA) using a Model TD-20/30 luminometer (Turner Designs). Light emission was measured for 10 s after injection. The specific luciferase activity of different transfections was determined in triplicate samples and was normalized on the Renilla luciferase activity. Data were expressed as means±s.d. of at least three independent experiments. Transient transfection efficiency in M31 and 2017, as assessed by expression of a GFP expression vector added in trace amounts, was ∼70 and ∼40%, respectively.
ChIP assay
Protein complexes were crosslinked to DNA in living nuclei of freshly isolated thymocytes to a final concentration of 1%. Crosslinking cells were processed, as previously described (Vacca et al, 2006) and divided into aliquots. In total, 5 μg of antibody (anti-RBP-Jk sc-28713X, anti-IK sc-13039, rabbit IgG sc-2027, Santa Cruz Biotechnology Inc.) was added to each aliquot of chromatin and incubated on a rotating platform for 12–16 h at 4°C. Antibody–protein–DNA complexes were isolated by immunoprecipitation with salmon sperm DNA/Protein A agarose (#157, Upstate Biotechnology). Following extensive washing, bound DNA fragments were eluted and analyzed by subsequent PCR using the following primers specific for the pTα mouse promoter including the CSL-core 1 site: 5′-GCTTTGGATCTGGAGGATGA-3′ and 5′-GAACTCAGGTCCCACTCCCA-3′.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Acknowledgments
We thank Massimo Zani for sequencing assistance. This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC), the Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR), the Ministero della Salute, the BEMM Center of Excellence and the ‘Eleonora Lorillard Spencer Cenci' Foundation.
References
- Akamatsu W, Fujihara H, Mitsuhashi T, Yano M, Shibata S, Hayakawa Y, Okano HJ, Sakakibara S, Takano H, Takano T, Takahashi T, Noda T, Okano H (2005) The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc Natl Acad Sci USA 102: 4625–4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allman D, Karnell FG, Punt JA, Bakkour S, Xu L, Myung P, Koretzky GA, Pui JC, Aster JC, Pear WS (2001) Separation of Notch1 promoted lineage commitment and expansion/transformation in developing T cells. J Exp Med 194: 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellavia D, Campese AF, Alesse E, Vacca A, Felli MP, Balestri A, Stoppacciaro A, Tiveron C, Tatangelo L, Giovarelli M, Gaetano C, Ruco L, Hoffman ES, Hayday AC, Lendahl U, Frati L, Gulino A, Screpanti I (2000) Constitutive activation of NF-kappaB and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J 19: 3337–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellavia D, Campese AF, Checquolo S, Balestri A, Biondi A, Cazzaniga G, Lendahl U, Fehling HJ, Hayday AC, Frati L, von Boehmer H, Gulino A, Screpanti I (2002) Combined expression of pTalpha and Notch3 in T cell leukemia identifies the requirement of preTCR for leukemogenesis. Proc Natl Acad Sci USA 99: 3788–3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverly LJ, Capobianco AJ (2003) Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell 3: 551–564 [DOI] [PubMed] [Google Scholar]
- Deschenes-Furry J, Belanger G, Perrone-Bizzozero N, Jasmin BJ (2003) Post transcriptional regulation of acetylcholinesterase mRNAs in nerve growth factor-treated PC12 cells by the RNA-binding protein HuD. J Biol Chem 278: 5710–5717 [DOI] [PubMed] [Google Scholar]
- Dumortier A, Jeannet R, Kirstetter P, Kleinmann E, Sellars M, Dos Santos NR, Thibault C, Barths J, Ghysdael J, Punt JA, Kastner P, Chan S (2006) Notch activation is an early and critical event during T-cell leukemogenesis in Ikaros-deficient mice. Mol Cell Biol 26: 209–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehling HJ, Iritani BM, Krotkova A, Forbush KA, Laplace C, Perlmutter RM, von Boehmer H (1997) Restoration of thymopoiesis in pTalpha−/− mice by anti-CD3epsilon antibody treatment or with transgenes encoding activated Lck or tailless pT alpha. Immunity 6: 703–714 [DOI] [PubMed] [Google Scholar]
- Fehling HJ, Krotkova A, Saint-Ruf C, von Boehmer H (1995) Crucial role of the pre-T-cell receptor alpha gene in development of alpha beta but not gamma delta T cells. Nature 375: 795–798 [DOI] [PubMed] [Google Scholar]
- Felli MP, Maroder M, Mitsiadis TA, Campese AF, Bellavia D, Vacca A, Mann RS, Frati L, Lendahl U, Gulino A, Screpanti I (1999) Expression pattern of notch1, 2 and 3 and Jagged1 and 2 in lymphoid and stromal thymus components: distinct ligand–receptor interactions in intrathymic T cell development. Int Immunol 11: 1017–1025 [DOI] [PubMed] [Google Scholar]
- Fryer CJ, Lamar E, Turbachova I, Kintner C, Jones KA (2002) Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev 16: 1397–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goolsby J, Marty MC, Heletz D, Chiappelli J, Tashko G, Yarnell D, Fishman PS, Dhib-Jalbut S, Bever CT Jr, Pessac B, Trisler D (2003) Hematopoietic progenitors express neural genes. Proc Natl Acad Sci USA 100: 14926–14931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm K, Ernst P, Lo K, Kim GS, Turck C, Smale ST (1994) The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol Cell Biol 14: 7111–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman MV, Levy S, Mock BA, Owens GC (1998) Gene organization and chromosome location of the neural-specific RNA binding protein Elavl4. Gene 208: 139–145 [DOI] [PubMed] [Google Scholar]
- Jeffries S, Robbins DJ, Capobianco AJ (2002) Characterization of a high-molecular-weight Notch complex in the nucleus of Notch(ic)-transformed RKE cells and in a human T-cell leukemia cell line. Mol Cell Biol 22: 3927–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehar SM, Dooley J, Farr AG, Bevan MJ (2005) Notch ligands Delta 1 and Jagged1 transmit distinct signals to T-cell precursors. Blood 105: 1440–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TD, Gao F, King PH, Andrews LG, Keene JD (1993) Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol 13: 3494–3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SE, Oyama T, Nagase T, Harigaya K, Kitagawa M (2002) Identification of new human mastermind proteins defines a family that consists of positive regulators for notch signaling. J Biol Chem 277: 50612–50620 [DOI] [PubMed] [Google Scholar]
- Lisbin MJ, Qiu J, White K (2001) The neuron-specific RNA-binding protein ELAV regulates neuroglian alternative splicing in neurons and binds directly to its pre-mRNA. Genes Dev 15: 2546–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobarak CD, Anderson KD, Morin M, Beckel-Mitchener A, Rogers SL, Furneaux H, King P, Perrone-Bizzozero NI (2000) The RNA-binding protein HuD is required for GAP-43 mRNA stability, GAP-43 gene expression, and PKC-dependent neurite outgrowth in PC12 cells. Mol Biol Cell 11: 3191–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Georgopoulos K (1994) The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol Cell Biol 14: 8292–8303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivero S, Maroc C, Beillard E, Gabert J, Nietfeld W, Chabannon C, Tonnelle C (2000) Detection of different Ikaros isoforms in human leukaemias using real-time quantitative polymerase chain reaction. Br J Haematol 110: 826–830 [DOI] [PubMed] [Google Scholar]
- Perrone-Bizzozero N, Bolognani F (2002) Role of HuD and other RNA-binding proteins in neural development and plasticity. J Neurosci Res 68: 121–126 [DOI] [PubMed] [Google Scholar]
- Primi D, Clynes RA, Jouvin-Marche E, Marolleau JP, Barbier E, Cazenave PA, Marcu KB (1988) Rearrangement and expression of T cell receptor and immunoglobulin loci in immortalized CD4−CD8− T cell lines. Eur J Immunol 18: 1101–1109 [DOI] [PubMed] [Google Scholar]
- Ratti A, Fallini C, Cova L, Fantozzi R, Calzarossa C, Zennaro E, Pascale A, Quattrone A, Silani V (2006) A role for the ELAV RNA-binding proteins in neural stem cells: stabilization of Msi1 mRNA. J Cell Sci 119: 1442–1452 [DOI] [PubMed] [Google Scholar]
- Reizis B, Leder P (2002) Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes Dev 16: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S, Natoli G (2002) Dynamic changes in histone H3 Lys 9 methylation occurring at tightly regulated inducible inflammatory genes. Genes Dev 16: 2219–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt TM, Zúñiga-Pflücker JC (2002) Induction of T cell development from hematopoietic progenitor cells by Delta-like-1 in vitro. Immunity 17: 749–756 [DOI] [PubMed] [Google Scholar]
- Spolski R, Miescher G, Erard F, Risser R, MacDonald HR, Mak TW (1988) Regulation of expression of T cell gamma chain, L3T4 and Ly-2 messages in Abelson/Moloney virus-transformed T cell lines. Eur J Immunol 18: 295–300 [DOI] [PubMed] [Google Scholar]
- Sun L, Crotty ML, Sensel M, Sather H, Navara C, Nachman J, Steinherz PJ, Gaynon PS, Seibel N, Mao C, Vassilev A, Reaman GH, Uckun FM (1999a) Expression of dominant-negative Ikaros isoforms in T-cell acute lymphoblastic leukemia1. Clin Cancer Res 5: 2112–2120 [PubMed] [Google Scholar]
- Sun L, Goodman PA, Wood CM, Crotty ML, Sensel M, Sather H, Navara C, Nachman J, Steinherz PG, Gaynon PS, Seibel N, Vassilev A, Juran BD, Reaman GH, Uckun FM (1999b) Expression of aberrantly spliced oncogenic ikaros isoforms in childhood acute lymphoblastic leukemia. J Clin Oncol 17: 3753–3766 [DOI] [PubMed] [Google Scholar]
- Sun L, Heerema N, Crotty L, Wu X, Navara C, Vassilev A, Sensel M, Reaman GH, Uckun FM (1999c) Expression of dominant-negative and mutant isoforms of the antileukemic transcription factor Ikaros in infant acute lymphoblastic leukemia. Proc Natl Acad Sci USA 96: 680–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talora C, Campese AF, Bellavia D, Pascucci M, Checquolo S, Groppioni M, Frati L, von Boehmer H, Gulino A, Screpanti I (2003) Pre-TCR-triggered ERK signalling-dependent downregulation of E2A activity in Notch3-induced T-cell lymphoma. EMBO Rep 4: 1067–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talora C, Sgroi DC, Crum CP, Dotto GP (2002) Specific down-modulation of Notch1 signaling in cervical cancer cells is required for sustained HPV-E6/E7 expression and late steps of malignant transformation. Genes Dev 16: 2252–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca A, Felli MP, Palermo R, Di Mario G, Calce A, Di Giovine M, Frati L, Gulino A, Screpanti I (2006) Notch3 and pre-TCR interaction unveils distinct NF-kappaB pathways in T-cell development and leukemia. EMBO J 25: 1000–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JPT, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC (2004) Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306: 269–271 [DOI] [PubMed] [Google Scholar]
- Winandy S, Wu L, Wang JH, Georgopoulos K (1999) Pre-T cell receptor (TCR) and TCR-controlled checkpoints in T cell differentiation are set by Ikaros. J Exp Med 190: 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winandy S, Wu P, Georgopoulos K (1995) A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell 83: 289–299 [DOI] [PubMed] [Google Scholar]
- Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD (2000) MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet 26: 484–489 [DOI] [PubMed] [Google Scholar]
- Zhu H, Hasman RA, Barron VA, Luo G, Lou H (2006) A nuclear function of Hu proteins as neuron-specific alternative RNA processing regulators. Mol Biol Cell 17: 5101 E-pub 2006 Oct 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3


