Abstract
We showed previously that eiger, the Drosophila tumor necrosis factor homolog, contributes to the pathology induced by infection with Salmonella typhimurium. We were curious whether eiger is always detrimental in the context of infection or if it plays a role in fighting some types of microbes. We challenged wild-type and eiger mutant flies with a collection of facultative intracellular and extracellular pathogens, including a fungus and Gram-positive and Gram-negative bacteria. The response of eiger mutants divided these microbes into two groups: eiger mutants are immunocompromised with respect to extracellular pathogens but show no change or reduced sensitivity to facultative intracellular pathogens. Hence, eiger helps fight infections but also can cause pathology. We propose that eiger activates the cellular immune response of the fly to aid clearance of extracellular pathogens. Intracellular pathogens, which can already defeat professional phagocytes, are unaffected by eiger.
Author Summary
We show that the gene eiger, which is the sole tumor necrosis factor homolog in the fruit fly, can play opposing roles in the fly's response to infections. Sometimes eiger contributes to the disease induced by an infection, while at other times it is required to fight an infection. Commonly, the fly's immune response is described as dividing microbes into two groups with Gram-positive bacteria and fungi lying in one group and Gram-negative bacteria lying in the other. Pathogenic bacteria can also be divided into two groups based on their behavior in eiger mutant flies, but these two groups differ from past descriptions. eiger tends to be required for the innate immune response against extracellular pathogens but tends to cause pathology during an infection with an intracellular pathogen. We suggest that eiger is required for innate immune responses that are effective at fighting extracellular pathogens but are wasteful or simply ineffective when fighting intracellular pathogens. We show here that the fly immune response is more complex than previously recognized and suggest new directions for studying pathogenesis in addition to innate immunity in the fly.
Introduction
The fruit fly has four main immune mechanisms to fight circulating microbes. These mechanisms include secreted antimicrobial peptides (AMPs), melanization, clotting, and phagocytic hemocytes [1−4]. The most deeply studied of these mechanisms, AMP secretion, is controlled by the Toll and imd pathways, which regulate the transcription of AMP genes in the fat body. Signaling through the Toll and imd pathways is activated by microbial elicitors; for example, the receptor peptidoglycan receptor protein LC, one receptor that triggers imd signaling, is activated by diaminopimelic acid–containing peptidoglycan [5,6]. This material is found on most Gram-negative bacteria as well as some Gram-positive bacteria.
Mutations affecting the Toll and imd pathways severely immunocompromise flies. Although Toll and imd mutant flies get sick and die from infections, this does not resemble many infectious processes in healthy humans. For example, an imd mutation essentially turns flies into a passive culture medium for Escherichia coli; the bacteria grow 1,000-fold in 24 h [7,8]. Immunocompromised infected flies likely die from the enormous load of bacteria that can reach levels of more than 1% of the mass of the fly. Death from massive numbers of microorganisms can happen in humans, particularly in immunocompromised patients; however, the infectious agents that are responsible for the greatest mortality in humans—Mycobacterium tuberculosis, HIV, Plasmodium falciparum, and diarrhea-inducing microbes—do not work in this manner. Instead, relatively small numbers of these infectious agents cause various pathologies that lead to death. To study microbial pathogenesis in the fly, it is necessary to follow microbes that cause disease in wild-type flies.
Pathogens are different from nonpathogenic bacteria and simple molecular elicitors of innate immunity, and thus the results we see from experiments with pathogens will be different than those observed for simple elicitors like E. coli and Micrococcus luteus. For one thing, pathogens can override the strong immune defenses of the fly; this causes disease. In flies, as in humans, there is more than one type of disease that results from infection. Pathogens have been observed to kill the fly in at least four different ways: first, overactivation of the Toll or imd pathways can be pathogenic [9,10]; second, Vibrio cholera fed to flies kills the fly through the secretion of toxins that presumably cause physiological changes to the gut [11]; third, M. marinum causes a wasting disease in flies [12]; and fourth, Salmonella typhimurium secretes effectors through its type III secretory system that increase the pathogenicity of the microbe. The fly gene eiger, the fly's sole tumor necrosis factor homolog, is implicated in causing pathology during this infection because S. typhimurium–infected eiger mutant flies live longer than infected wild-type flies [13]. It appears that there are many physiological routes that can lead to death following infection.
We have begun to try to understand these physiological routes to death by analyzing the role played by eiger in a variety of infections. The eiger mutation divided our group of pathogens into two unanticipated groups. The first group of microbes kills eiger mutants more rapidly than wild-type flies; this group includes a fungus as well as both Gram-positive and Gram-negative bacterial species. The second group of microbes kills eiger mutants at the same rate or more slowly than wild-type flies; this group includes a Gram-positive and a Gram-negative bacterial species and a mycobacterium.
These results cut across the previous descriptions of Drosophila immunity, which groups microbes according to their gross physical characteristics [2,3]. The most obvious way to explain the grouping of these pathogens in eiger mutants is not by grouping microbes according to their Gram-staining properties or kingdom, but rather by their pathogenesis mechanisms; the first group of microbes consists of extracellular pathogens while the second group contains microbes that can grow within professional phagocytes. eiger appears to play a role in innate immunity in fighting extracellular pathogens but plays a role in driving pathogenesis when fighting some intracellular pathogens. This study demonstrates that the examination of real pathogens and the use of outputs other than AMP transcription leads to the discovery of unanticipated immune pathways and reveals new complexities in the Drosophila immune system.
Results/Discussion
To determine the role eiger plays in fighting microbial pathogens, we tested a collection of Gram-positive, Gram-negative, and fungal pathogens that could cause either intracellular or extracellular infections (Table 1). Our goal was to test a broad group of microbes that used different virulence mechanisms and were recognized by different innate immunity pathways.
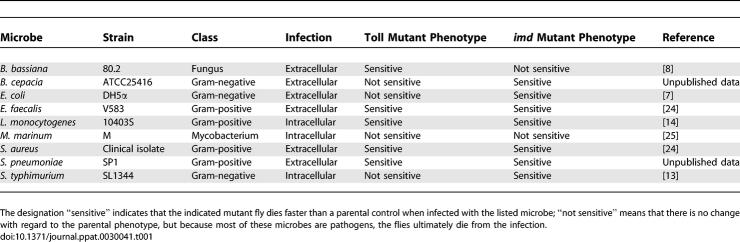
Table 1.
Microbial Strain List
We tested a heterozygous combination of two eiger null alleles (w1118; egr1/egr3) and compared this to an isogenic w1118 parental strain (Figure 1). eiger mutants died much faster than wild-type flies when challenged with Beauveria bassiana, Burkholderia cepacia, Enterococcus faecalis, Staphylococcus aureus, and Streptococcus pneumoniae (Figure 1). The members of this group of microorganisms are very different from each other and include Gram-positive and Gram-negative bacteria and a fungus. The smallest effect on survival was a 50% reduction in the mean time of death for B. bassiana infections. The effect was strongest with eiger mutants infected with S. aureus and E. faecalis; whereas the chosen doses were nonpathogenic to wild-type flies (i.e., they died at the same rate as media-injected controls), eiger mutants died within 2 d. The one common characteristic of this group is that these microbes are all expected to produce extracellular infections.
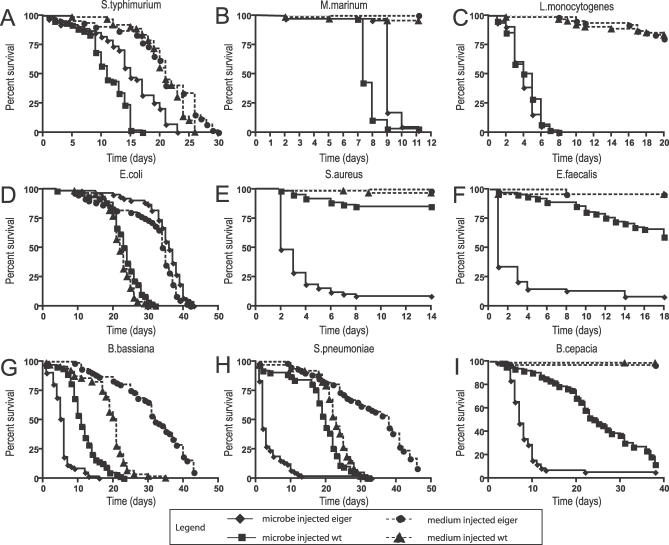
Figure 1. Survival of eiger Infected Flies.
Week-old male flies were infected with pathogens and survival was monitored daily. (A) S. typhimurium; (B) M. marinum, (C) L. monocytogenes, (D) E. coli, (E) S. aureus, (F) E. faecalis, (G) B. bassiana, (H) S. pneumoniae, (I) B. cepacia. Circles indicate medium-injected eiger; diamonds, microbe-injected eiger; squares, microbe-injected parental; triangles, medium-injected parental. Medium injection is indicated by a dotted line, while microbe injection is indicated by a solid line. Statistical significance was determined using log-rank analysis. The infected eiger and wild-type curves in (A), (B), and (E–I) are significantly different with p < 0.0001 as determined by log-rank analysis.
eiger mutant flies did not die faster when infected with the three facultative intracellular pathogens we tested (Figure 1). We reported previously that eiger mutants live longer than wild-type flies when infected with S. typhimurium. This result led us to suggest that eiger activity could be deleterious for the fly and was a cause of pathology. We found that M. marinum–infected eiger mutants also live longer than infected wild-type flies. Finally, eiger mutants and wild-type flies survive exactly the same amount of time when infected with Listeria monocytogenes.
We tested E. coli as a nonpathogenic control (Figures 1 and S1). We define a pathogen as a microbe that increases the death rate of infected flies as compared to a control fly injected with medium. This microbe was chosen because it played a major role in characterizing the immune response of the fly. E. coli is normally a good inducer of innate immune responses when injected into the fly but will only cause disease in flies missing the imd pathway. E. coli injection does not kill eiger mutants or wild-type flies. This suggests either that eiger does not play a role in fighting a nonpathogenic infection or that eiger's role is redundant and therefore undetectable by this assay.
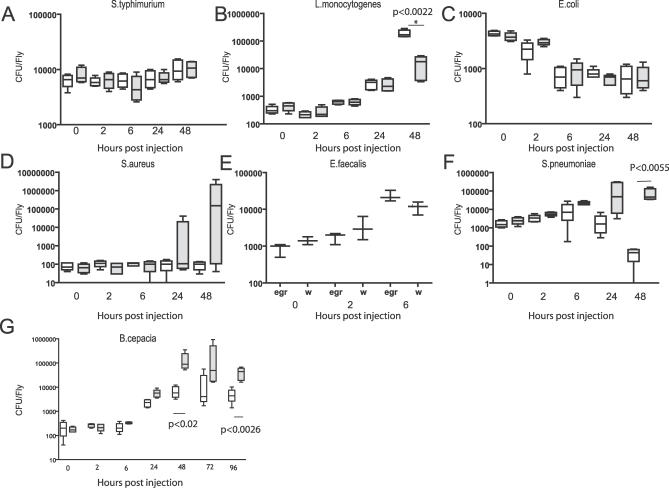
We next decided to determine whether eiger mutant flies were killed faster by extracellular pathogens because of increased bacterial growth or increased pathogenesis by monitoring bacterial proliferation in infected flies (Figure 2). As before, bacteria were injected into age-matched male wild-type parental or isogenic transheterozygous eiger mutant flies. Flies were collected and homogenized following bacterial challenge to measure bacterial loads.
Figure 2. Bacterial Growth in eiger Mutants.
Week-old male flies were infected with pathogens, and flies were collected at 0, 2, 6, 24, and 48 h, if there were survivors. Live flies were homogenized and plated: (A) S. typhimurium; (B) L. monocytogenes; (C) E. coli; (D) S. aureus; (E) E. faecalis; (F) S. pneumoniae; (G) B. cepacia. Data are plotted as box plots with whiskers. White bars indicate the parental w1118 line; gray bars, w1118; eiger1/eiger3 mutants. Statistical significance was calculated using two-tailed nonparametric t-tests.
Of the four bacterial species that showed increased virulence in eiger mutants, two—B. cepacia and S. pneumoniae—had statistically significant increased growth rates in eiger mutants. There was no clear effect on E. faecalis growth rates in eiger versus wild-type flies. S. aureus showed a large variation in bacterial numbers toward the end of the infection that suggested a trend toward increased growth; for example, the highest levels of bacteria found in eiger mutants were 10,000 higher than those seen in wild-type flies. However, because the variation was so great in eiger mutants, the difference between wild-type and eiger mutants was not statistically significant. We suggest that S. aureus might not cause synchronous infections and thus that some flies succumb to infection rapidly and have high numbers of bacteria while others take longer to die and maintain lower levels of bacteria. This would result in a huge range of bacterial titers in a group of injected flies. E. coli were cleared at comparable rates in eiger and wild-type flies. B. bassiana was not tested because we do not have a good quantitative method of measuring fungal growth. In summary, these bacterial growth measurements show that eiger mutants are unable to limit the growth of two and possibly three of the four extracellular pathogens tested. This experiment does not rule out the possibility that eiger mutants suffer greater pathological effects of infection but does suggest that eiger in a wild-type fly plays a role in reducing the numbers of extracellular microbes.
The proliferation of facultative intracellular pathogens did not increase in eiger mutants. As we published previously, S. typhimurium levels remained constant in an eiger homozygote as compared to an isogenic wild-type fly [13]. L. monocytogenes numbers decreased significantly in eiger mutants with respect to wild-type flies even though both wild-type and mutant flies died at the same rate. Our interpretation is that eiger is somehow helpful for the growth of Listeria. We did not measure the growth rate of M. marinum.
There is no simple interpretation, using Toll or imd signaling, that can explain the difference in sensitivity of eiger mutants to the tested pathogens. If the eiger mutation resulted in decreased Toll signaling, then we would expect Listeria to kill the flies rapidly as has been reported [14]. Likewise, a reduction of imd activity would immunocompromise the flies to S. typhimurium. imd mutants are very sensitive to S. typhimurium and will die within 24 h when infected with as little as a single colony-forming unit [13]. Furthermore, if imd signaling was affected, we would expect flies to become sensitive to E. coli, which they do not [7]. We are forced to conclude that the strong effects of eiger on the pathogenesis caused by extracellular microbes are not caused by a reduction in signaling through the Toll and imd pathways.
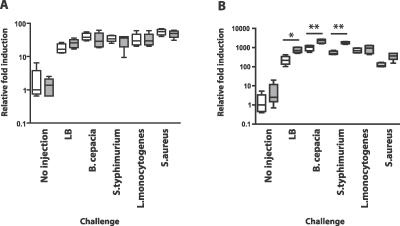
To test the hypothesis that Toll and imd signaling are not grossly reduced by a mutation in eiger, we measured the induction of AMP transcription in eiger and parent strains (Figure 3). Flies were challenged with S. typhimurium, B. cepacia, L. monocytogenes, and S. aureus as well as an LB control, and these conditions were compared to expression levels seen in uninfected flies. We chose this subset of bacteria because it included both Gram-positive and Gram-negative examples of intracellular and extracellular pathogens. Toll signaling was monitored by following drosomycin gene induction using quantitative reverse transcription (RT)-PCR, although we note that there are no perfect Toll responsive genes and drosomycin is responsive to both Toll and imd signaling [15]. To measure the output of the imd pathway, we monitored the expression of the diptericin gene. No statistically significant changes in drosomycin transcription were seen between parental and eiger mutant flies. Some conditions caused a relatively small, but statistically significant increase in diptericin transcription in eiger mutants. We do not understand how the slightly higher levels of AMP transcription found in eiger mutants might affect the eiger phenotype. The important point is that eiger mutants did not have lower levels of AMP gene expression than did wild-type flies, demonstrating that the eiger phenotype is not caused by reduction in Toll or imd signaling.
Figure 3. Antimicrobial Gene Expression Differences between eiger Mutants and Wild-Type Controls.
Flies were injected with an LB control, B. cepacia, S. typhimurium, L. monocytogenes, or S. aureus or left uninjected. RNA was harvested after a 6-h incubation at 29 °C, and quantitative RT-PCR was used to assay drosomycin (A) and diptericin (B) gene expression levels relative to a ribosomal protein 15a control. White bars indicate the w1118 parental control; grey bars, w1118; egr1/egr3 One asterisk indicates p < 0.01; two asterisks, p < 0.001 using Tukey's multiple-comparison test following one-way ANOVA. Unmarked parental/mutant pairs do not differ in a statistically significant manner. No differences were seen with drosmycin expression between mutant and parental lines, whereas eiger mutant flies transcribed 2- to 3-fold more diptericin (during a 150- to 1,100-fold induction compared with uninjected flies) than did their parents, when challenged with LB, B. cepacia, or S. typhimurium.
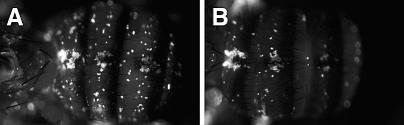
As Toll and imd signaling, and thus the majority of the antimicrobial peptide response in the fly [15], does not seem to be responsible for changes in eiger mutants, we next probed the cellular immune response (Figure 4). To determine whether eiger mutations might affect hemocyte function, we monitored phagocytosis in eiger mutants. Fluorescein isothiocyanate (FITC)-labeled S. aureus were injected into the hemocoel of wild-type or eiger mutant flies and the injected flies were given 1 h to phagocytose the particles. S. aureus was chosen because FITC-labeled dead S. aureus are commercially available and we predicted that eiger alters the ability of hemocytes to fight this pathogen. Trypan blue was injected into the hemocoel of the flies following the 1-h incubation period. This dye quenches the fluorescence of extracellular FITC but allows phagocytosed particles to fluoresce brightly [16]. The flies were examined around the dorsal anterior abdomen because hemocytes tend to gather in this area. Changes in phagocytosis can be seen by changes in the numbers of phagocytosing cells or by the fluorescence intensity. Both of these characteristics appear altered in eiger mutants compared to parental flies, suggesting that eiger mutant hemocytes were either reduced in number or had reduced phagocytic activity, or both.
Figure 4. Phagocytosis in eiger Mutants.
Week-old male flies were injected with FITC-labeled dead S. aureus and allowed to phagocytose the particles and then were injected with Trypan blue to quench extracellular fluorescence. The dorsal abdomen of the fly was photographed under epifluoresence optics. Anterior is on the left. (A) Wild-type fly (w1118); (B) eiger mutant fly (w1118; egr1/egr3). Bright spots indicate hemocytes that have phagocytosed the S. aureus. The pictures are representative of the 15 flies examined for the experiment.
We note that uninfected eiger mutants live longer than wild-type flies. This raises the possibility that the increased survival of eiger mutant flies after infection with M. marinum or S. typhimurium is simply a consequence of general increased longevity. We argue that this possibility is unlikely, because it assumes that the cause of death due to old age is the same as the cause of death by infection; this is not observed in other animals and it is not supported by examination of published microarray studies of flies dying from infection or old age [12,17,18]. Instead, we argue that eiger is a driver of pathogenesis. If eiger is indeed required to fight infections but can also cause pathology, then the phenotype we observed for a given infection is likely the sum of the positive and negative effects of eiger. Regardless of the interpretation of the effects of eiger on S. typhimurium and M. marinum, the important result remains that eiger mutations divide the fly's response to pathogens into two groups.
Why does eiger affect different microbes in different ways? We argue that E. coli is removed from flies so rapidly and via so many mechanisms that the effects of eiger cannot be measured easily because of redundancy. In contrast, during an infection with a real pathogen, it may be easier to measure changes in immunity because the fly is fighting hard for survival and its immune mechanisms are not acting in a redundant fashion. Extracellular pathogens are clearly fought by the fly using eiger-dependent mechanisms because the loss of eiger results in a deeply sensitive phenotype. It is intriguing that flies lacking eiger are no worse at fighting intracellular pathogens. This suggests that these intracellular pathogens are normally immune to the effects of wild-type eiger. Intracellular pathogens like M. marinum, L. monocytogenes, and S. typhimurium use different virulence mechanisms for growing inside cells, but the common thread is that they can survive in professional phagocytes. We suggest that eiger function somehow alters hemocytes to increase their potency against microbes. The reduced ability of eiger mutant hemocytes to phagocytose S. aureus compared to wild-type hemocytes supports this hypothesis. We predict that this change in potency is effective against pathogens that grow extracellularly but not against pathogens that have already developed methods of defeating phagocytes.
We propose that where eiger signaling does not help fight infection, eiger can cause pathology. This eiger-induced pathology may be linked to the immune function of eiger—for example, the induced immune response may be energetically wasteful or directly toxic. Alternatively, eiger-induced pathology may be separable from the immune function—for example, eiger could cause something like muscle wasting in the fly, as has been suggested for tumor necrosis factor in vertebrates [19].
Genetic screens that monitored AMP synthesis have been productive and filled in the Toll and imd pathways but did not reveal eiger signaling [20,21]. As we have shown, the role eiger plays in innate immunity cannot be measured using a nonpathogenic microbe like E. coli. This study demonstrates that there are important immune mechanisms at work in the fly that are difficult to see using simple endpoints like antimicrobial gene expression; however, studies with microbes that can cause disease in wild-type flies—real pathogens—can reveal these physiologies.
Materials and Methods
Fly strains.
The wild-type parental strain used in all experiments is white1118. The eiger alleles white1118;egr1 and white1118;egr3 were kept as homozygous stocks and crossed to make heterozygous flies as required [22]. We used this heterozygous combination to reduce the probability that there were other mutations on the chromosome that affected pathogenesis. To control for variation in the flies, we infected only 5- to 7-d-old male flies.
Bacterial strains.
Strains used are listed in Table 1.
Pathogen culture conditions.
B. bassiana cultures were grown on malt agar at 29 °C for 2 wk or until a sufficient density was reached, and the cultures were allowed to sporulate. Anesthetized flies were shaken on the plates for 30 s to coat flies with spores. Flies were transferred to fresh vials and incubated at 29 °C for the duration of the survival experiment.
S. pneumoniae cultures were grown standing at 37 °C 5% CO2 in brain heart infusion medium (BHI) to an OD600 of 0.15, and aliquots were frozen at −80 °C in 10% glycerol. For infection, an aliquot of S. pneumoniae was thawed, diluted 1:3 in fresh BHI, and allowed to adjust at 37 °C 5% CO2 for 1.5 h. E. coli, S. typhimurium, E. faecalis, and S. aureus cultures were grown overnight in Luria Bertani (LB) medium at 37 °C. L. monocytogenes was grown standing overnight in BHI medium.B. cepacia, L. monocytogenes, S. pneumoniae, and S. typhimurium were grown standing, while E. coli, E. faecalis, and S. aureus were shaken. M. marinum was cultured standing at 29 °C in Middlebrook 7H9 broth supplemented with Middlebrook oleic acid–albumin–dextrose–catalase enrichment and 0.2% Tween. Then 50 nl of bacteria was injected at the following optical densities (OD600): B. cepacia, 0.0001–0.001;E. coli, 0.1; E. faecalis, 0.5; L. monocytogenes, 0.01; M. marinum, 0.05; S. aureus, 0.001; S. pneumoniae, 0.05; and S. typhimurium, 0.1.
Injection.
Five- to 7-d posteclosion male flies were used for injection. The flies were raised at 25 °C, 65% humidity, on yeasted dextrose food. Flies were anesthetized with CO2. Injections were carried out with a pulled glass capillary needle. A picospritzer (Parker Hannifin, http://www.parker.com) was used to inject 50 nl of liquid into each fly with needles that were individually calibrated by measuring the size of the expelled drop under oil. Reproducibility was measured by determining the number of bacteria injected at time zero and can be seen in Figure 2. Injected flies were incubated 20 flies to a vial and placed at 29 °C, 65% humidity with the exception of B. cepacia and M. marinum. B. cepacia infections were performed at 18 °C in the dark, and humidity was not controlled in this experiment. This temperature was used because B. cepacia is so pathogenic that it is difficult to obtain sufficient survival at higher temperatures to observe changes in death rates. M. marinum infections were carried out at 25 °C, 65% humidity, in the dark.
Survival curves.
Parental flies (w1118) and w1118; eiger1/eiger3 mutants were injected with the microbe of choice or medium as a control. Sixty flies were assayed for each survival curve, and they were placed in three vials of 20 flies each. Death was recorded daily. Data were not censored. Survival curves are plotted as Kaplan-Meier plots, and statistical significance is tested using log-rank analysis using Prism software (http://www.prism-software.com). Kaplan-Meier plots are shown in Figure 1, and survival curves showing the variance in the data are included in Figure S1. All experiments were performed at least three times and yielded similar results.
CFU determination.
Following challenge with microbes, six individual flies were collected at each time point. These flies were homogenized, diluted serially, and plated onto appropriate media (blood agar for S. pneumoniae, LB for all others). E. faecalis CFUs were determined by testing three groups of six flies for each time point. The data are plotted as boxes with whiskers. The median is indicated with a bold line. The boxes indicate the extent of the third and first quartiles, while the whiskers show the complete range of the data. Statistical significance was determined using nonparametric two-tailed t-tests. All experiments were performed at least three times and yielded similar results.
Antimicrobial peptide gene expression.
Flies were injected with 50 nl of the indicated microbes or controls. Following injection, the flies were placed in vials containing yeasted dextrose food and incubated at 29 °C for 6 h. Groups of five flies were homogenized in TriZOL and stored at −70 °C until processed. RNA was isolated using a standard TriZOL preparation, and the samples were treated with DNase (Promega, http://www.promega.com). Quantitative RT-PCR was performed as described [23] previously using a Bio-Rad icycler (http://www.bio-rad.com) and the following primer sets: drosomycin 5′-gracttgttcgccctcttcg, drosomycin 3′-cttgcacacacgacgacag, drosomycin Taqman probe tccggaagatacaagggtccctgtg, diptericin 5′-accgcagtacccactcaatc, diptericin 3′-cccaagtgctgtccatatcc, and diptericin Taqman probe cagtccagggtcaccagaaggtgtg. The data shown in Figure 4 include six biological replicates of each treatment condition, and each data point was calculated as the mean of two technical replicates. This experiment was repeated once using a set of three biological replicates with similar results. The data are plotted as boxes with whiskers. The median is indicated with a bold line. The boxes indicate the extent of the third and first quartiles, while the whiskers show the complete range of the data.
Phagocytosis assays.
Flies were injected with 50 nl of 1 mg/ml FITC-labeled S. aureus (Molecular Probes, http://probes.invitrogen.com) in water. The flies were allowed to phagocytose the particles for 1 h and then were injected in the thorax with approximately 250 μl of 4% Trypan blue: this quenches the fluorescence of extracellular bacteria but permits the phagocytosed particles to fluoresce. The wings of the flies were removed with iris scissors, and the flies were pinned with a minuten pin and photographed using epifluorescent illumination with a Leica MZ3 microscope fitted with an ORCA camera. Images were captured with Openlab (Improvision) software (http://www.improvision.com). The exposure was set such that the brightest images had a small number of saturating pixels. The experiment was repeated three times with at least five flies for each treatment.
Supporting Information
Week-old male flies were infected with pathogens and survival was monitored daily. (A) S. typhimurium; (B) M. marinum, (C) L. monocytogenes, (D) E. coli, (E) S. aureus, (F) E. faecalis, (G) B. bassiana, (H) S. pneumoniae, and (I) B. cepacia. Circles indicate medium-injected eiger; diamonds, microbe-injected eiger; squares, microbe-injected parental; triangles, medium-injected parental. The mean is plotted, and error bars show the standard deviation from three groups of 20 injected flies.
(1.4 MB AI).
Acknowledgments
We thank E. J. Baron, S. Falkow, N. Silverman, and J. Theriot for microbial strains. We thank M. Miura for the eiger mutants and the parental line.
Abbreviations
- AMP
antimicrobial peptide
- FITC
fluorescein isothiocyanate
- RT
reverse transcription
Footnotes
¤ Current address: Departments of Craniofacial Development and Microbiology, King's College London, London, United Kingdom
Competing interests. The authors have declared that no competing interests exist.
Author contributions. JSA, SMB, AC, MSD, MDG, EMM, MGM, LNP, and MMSH contributed equally to this paper. DSS, JSA, SMB, AC, MSD, MDG, EMM, MGM, LNP, and MMSH conceived and designed the experiments, performed the experiments, and wrote the paper. DSS, MSD, MDG, EMM, MGM, LNP, and MMSH analyzed the data. JSA, SMB, and AC contributed reagents/materials/analysis tools.
Funding. This work was supported by National Institutes of Health grants AI053080 and AI055651.
References
- Bidla G, Lindgren M, Theopold U, Dushay MS. Hemolymph coagulation and phenoloxidase in Drosophila larvae. Dev Comp Immunol. 2005;29:669–679. doi: 10.1016/j.dci.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Brennan CA, Anderson KV. Drosophila: The genetics of innate immune recognition and response. Annu Rev Immunol. 2004;22:457–483. doi: 10.1146/annurev.immunol.22.012703.104626. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA. The immune response of Drosophila . Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- Meister M, Lagueux M. Drosophila blood cells. Cell Microbiol. 2003;5:573–580. doi: 10.1046/j.1462-5822.2003.00302.x. [DOI] [PubMed] [Google Scholar]
- Stenbak CR, Ryu JH, Leulier F, Pili-Floury S, Parquet C, et al. Peptidoglycan molecular requirements allowing detection by the Drosophila immune deficiency pathway. J Immunol. 2004;173:7339–7348. doi: 10.4049/jimmunol.173.12.7339. [DOI] [PubMed] [Google Scholar]
- Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, et al. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol. 2003;4:478–484. doi: 10.1038/ni922. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, et al. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc Natl Acad Sci U S A. 1995;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Nicolas E, Michaut L, Reichhart J, Hoffmann J. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, et al. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2006;2:e14. doi: 10.1371/journal.ppat.0020014. doi: 10.1371/journal.ppat.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MD, Dionne MS, Schneider DS, Nusse R. WntD is a feedback inhibitor of Dorsal/NF-kappaB in Drosophila development and immunity. Nature. 2005;437:746–749. doi: 10.1038/nature04073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watnick TJ, Jin Y, Matunis E, Kernan MJ, Montell C. A flagellar polycystin-2 homolog required for male fertility in Drosophila . Curr Biol. 2003;13:2179–2184. doi: 10.1016/j.cub.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Dionne MS, Pham LN, Shirazu-Hiza MM, Schneider DS. Akt and FOXO dysregulation contribute to infection-induced wasting in Drosophila . Curr Biol. 2006;16:1977–1985. doi: 10.1016/j.cub.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Brandt SM, Dionne MS, Khush RS, Pham LN, Vigdal TJ, et al. Secreted bacterial effectors and host-produced eiger/TNF drive death in a Salmonella-infected fruit fly. PLoS Biol. 2004;2:e418. doi: 10.1371/journal.pbio.0020418. doi: 10.1371/journal.pbio.0020418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield BE, Dionne MS, Schneider DS, Freitag NE. Exploration of host-pathogen interactions using Listeria monocytogenes and Drosophila melanogaster . Cell Microbiol. 2003;5:901–911. doi: 10.1046/j.1462-5822.2003.00329.x. [DOI] [PubMed] [Google Scholar]
- De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila . EMBO J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrod-Erickson M, Mishra S, Schneider D. Interactions between the cellular and humoral immune responses in Drosophila . Curr Biol. 2000;10:781–784. doi: 10.1016/s0960-9822(00)00569-8. [DOI] [PubMed] [Google Scholar]
- Landis GN, Abdueva D, Skvortsov D, Yang J, Rabin BE, et al. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster . Proc Natl Acad Sci U S A. 2004;101:7663–7668. doi: 10.1073/pnas.0307605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Meadows S, Sharp L, Jan LY, Jan YN. Genome-wide study of aging and oxidative stress response in Drosophila melanogaster . Proc Natl Acad Sci U S A. 2000;97:13726–13731. doi: 10.1073/pnas.260496697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman RW, Kandarian SC. The molecular basis of skeletal muscle atrophy. Am J Physiol Cell Physiol. 2004;287:C834–C843. doi: 10.1152/ajpcell.00579.2003. [DOI] [PubMed] [Google Scholar]
- Foley E, O'Farrell PH. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol. 2004;2:e203. doi: 10.1371/journal.pbio.0020203. doi: 10.1371/journal.pbio.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LP, Choe KM, Lu Y, Anderson KV. Drosophila immunity: Genes on the third chromosome required for the response to bacterial infection. Genetics. 2001;159:189–199. doi: 10.1093/genetics/159.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaki T, Kanda H, Yamamoto-Goto Y, Kanuka H, Kuranaga E, et al. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J. 2002;21:3009–3018. doi: 10.1093/emboj/cdf306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Shahabuddin M. Malaria parasite development in a Drosophila model. Science. 2000;288:2376–2379. doi: 10.1126/science.288.5475.2376. [DOI] [PubMed] [Google Scholar]
- Pili-Floury S, Leulier F, Takahashi K, Saigo K, Samain E, et al. In vivo RNA interference analysis reveals an unexpected role for GNBP1 in the defense against Gram-positive bacterial infection in Drosophila adults. J Biol Chem. 2004;279:12848–12853. doi: 10.1074/jbc.M313324200. [DOI] [PubMed] [Google Scholar]
- Dionne MS, Ghori N, Schneider DS. Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum . Infect Immun. 2003;71:3540–3550. doi: 10.1128/IAI.71.6.3540-3550.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Week-old male flies were infected with pathogens and survival was monitored daily. (A) S. typhimurium; (B) M. marinum, (C) L. monocytogenes, (D) E. coli, (E) S. aureus, (F) E. faecalis, (G) B. bassiana, (H) S. pneumoniae, and (I) B. cepacia. Circles indicate medium-injected eiger; diamonds, microbe-injected eiger; squares, microbe-injected parental; triangles, medium-injected parental. The mean is plotted, and error bars show the standard deviation from three groups of 20 injected flies.
(1.4 MB AI).