Abstract
The human telomerase reverse transcriptase (hTERT) is the catalytic subunit of the enzyme telomerase which is responsible for telomeric maintenance and extension. Using RNA interference to knock down hTERT mRNA expression, we provide evidence that hTERT exerts extra-telomeric effects on the cell cycle and on its own regulatory proteins, specifically: p53 and p21. We tested our hypothesis that hTERT regulates its own expression through effects on upstream regulatory genes using transformed human embryonic kidney (HEK 293) cells, p53 and p16INK4a null human ovarian cancer SKOV-3 cells, and p53-null MDA-MB-157 human mammary cancer cells. In HEK 293 cells, hTERT knock down resulted in elevated p53 and p21 transcription and a decrease in cellular proliferation. Similar results were observed in the MDA-MB-157 cell line where p21 was up-regulated, correlating with cell growth inhibition. In contrast, we observed a decrease in expression of p21 in SKOV-3 cells with hTERT knock down and cell growth appeared to be unaffected. These findings suggest that hTERT may be involved in a feedback loop system, thereby playing a role in its own regulation.
Keywords: Telomerase, Telomere, hTERT, Cell cycle, p53, p21
Introduction
Telomerase is a DNA polymerase that uses an RNA template in a reverse transcription process to synthesize telomeric ends on linear chromosomes during replication to counteract the “end-replication problem”. Telomerase activity is barely detectable in somatic cells but is reactivated in immortalized cell lines and human cancers [1, 2]. Due to this repression of telomerase, normal somatic cells experience telomeric attrition at a mean loss of 30-150 bps of telomeric DNA with each cell replication [3] until a critical minimum telomeric length is reached, at which time the cells experience cellular senescence [4]. Telomeres serve to preserve chromosomal integrity by preventing rearrangements, nuclease degradation, and end-to-end chromosomal fusions [5, 6]. The telomerase holoenzyme is a ribonucleoprotein complex; although it is comprised of many components, only two subunits are essential to its function. In humans, the catalytic subunit of telomerase, hTERT, confers its reverse transcriptase activity, adding 5’-TTAGGG-3’ repeat sequences onto telomeres; the RNA component, hTR, is complementary to the telomeric repeat sequence, serving as a template for elongation of the telomeres [7-9]. The hTR component of telomerase is found to be ubiquitously expressed in most cell types including telomerase-negative cells, such as differentiated somatic cells [10, 11]. hTERT, on the other hand, is tightly regulated during differentiation and is almost undetectable in most somatic cells [12]. A positive correlation has been found between the amount of mRNA of hTERT and the activity of telomerase, suggesting that telomerase activity is regulated at the gene transcriptional level of hTERT [13-16]. Therefore, inhibition of hTERT expression usually results in decreasing of telomeric length.
For the more than 20 years since Carol Greider discovered telomerase [7], scientists have believed that telomerase is a terminal effector and that its role is mainly to extend and maintain the length of telomeres. However, recent studies have shown that the expression of hTERT affects a variety of other genes. In a recent study in which microarray analysis of 19,000 genes was performed in bovine adrenocortical cells ectopically expressing hTERT, 284 genes were found to be either positively or negatively affected by changes in hTERT, suggesting that hTERT may have extra-telomeric effects. Interestingly, most of the genes affected were involved in cell-cycle regulation, signaling, and metabolism [17]. Further, overexpression of telomerase due to the ectopic expression also resulted in a decrease in the mRNA of tp53bp1, a gene involved in the induction of the p53/p21 system [17]. In this study, we hypothesize that hTERT exerts its effects not only on telomeres and other genes, but also specifically on its upstream regulators, mainly p53 and p21, thereby partially regulating itself.
p53 is a tumor suppressor protein that functions in differentiation, apoptosis, DNA repair and control of cell cycle progression, by regulating the transcription of many genes, both negatively and positively. The hTERT gene has two p53 binding motifs at -1877 and -1240 relative to the start of transcription, upstream of the 5’ core promoter region (18). Overexpression of p53 and binding of the protein with the assistance of transcription factor Sp1 at these two motifs represses the hTERT promoter [18]. In mammary epithelial cells, abrogation of p53 function induces cellular immortality, probably through the reactivation of telomerase [18].
The p21WAF1/Cip1/SDI1 (p21) gene is ubiquitously expressed in mammalian cells, and as a major player in cell cycle arrest, is critical for the control of differentiation, senescence, and apoptosis [19, 20]. Over-expression of p21 induces a correlative repression of hTERT, which has been demonstrated in human glioma cells [21] and in squamous cell carcinoma where telomerase expression is abnormally elevated [22]. p21 contains a p53 responsive element at -2281/-2262 in the distal region of its promoter. Full transactivation of the p21 promoter is enabled by the binding of p53 protein, Sp1 proteins and related factors at the promoter region [23, 24]. The downstream effect of p53 transactivation of the p21 promoter is the negative regulation of the cell cycle, thereby preventing progression past the G1/S checkpoint [25]. At the same time, p21 activation leads to the repression of the hTERT promoter, thereby effectively shutting down the cell cycle, as we will show in this study.
We analyzed human embryonic kidney (HEK 293), human ovarian cancer (SKOV-3), and human mammary cancer (MDA-MB-157) cell lines, to assess the effects of hTERT knock down the levels of p53 and p21, and the subsequent effect on cell proliferation. HEK cells are adenovirally transformed and express adenoviral E1A and E1B proteins [26]. A transformed cell line was chosen to show the effects of hTERT knock down on the proliferation of transformed cells and to investigate the potential of RNA interference (RNAi) of hTERT for therapeutic purposes. Also, because these cells are embryonic in origin, proliferate indefinitely in vitro and do not show contact inhibition, they were appropriate for the short-term dsRNA knock down of hTERT and for proliferation studies. Further, studies have indicated that mammalian cells of embryonic origin lack the interferon response upon treatment with double-stranded RNA [27-29]. Adenoviral E1B and E1A proteins inactivate p53 either by physically binding p53 or by binding its downstream effectors such as mdm-2 and other apoptotic components [30-32]. However, adenovirally transformed cells can accumulate p53 to an extent where excess p53 is transcriptionally active [33]. Thus, HEK cells are not G1-arrested, have high levels of telomerase [34], and have transcriptionally active p53 [33].
P53 and p16INK4a null SKOV-3, and p53-null MDA-MB-157 cell lines [35] were utilized in this study as p53 deficient model systems to determine the effects of long-term hTERT knock down on p21 levels and cell proliferation. We demonstrate how hTERT levels can directly affect p21, since the mechanism of p53-dependent transactivation of p21 is eliminated in these cell lines.
Using specific RNAi knock down of hTERT in a variety of different cell types, our studies collectively demonstrate for the first time that the level of hTERT in the cell may regulate the expression of its gene.
Materials and methods
dsRNA Synthesis
A DNA template for the synthesis of dsRNA was first generated by PCR amplifying a 219 bp fragment of HEK cDNA specific to the hTERT gene transcript with primers: ElmoreF (5’-GACTCGACACCGTGTTCACCTAC-3’) and ElmoreR (5’-ACGTAGAGCCCGGCGTGACAG-3’), each constructed with T7 promoter sequences attached to their 5’-ends (5’-TAATACGACTCACTATAGGGAGA-3’) as indicated by Ambion MEGAscript RNAi kit. dsRNA was generated using MEGAscript RNAi Kit [Ambion, Austin, TX], according to the manufacturer’s instructions. The generated single-stranded RNAs were heated to 75°C for 5 min and allowed to slowly anneal at room temperature until cooled.
Plasmid Construct
pSilencer 3.1 H1-neo [Ambion Inc.] plasmid was digested with restriction enzymes BamHI and HindIII at 37°C for 1 h and the enzymes were inactivated at 65°C for 15 min. Oligonucleotide fragments were removed from the reaction using the Qiaquick DNA Cleanup Systems, Nucleotide Removal protocol [Qiagen, Valencia, CA]. A plasmid construct, pS2-3, targeting the upstream section of hTERT mRNA beginning at +863, was used to determine the effects of long-term hTERT knockdown. The insert sequence for pS2-3 is as follows: Sense: 5’-GATCCGTTCTGTGTGGTGTCACCTGTTCAAGAGACAGGTGACACCACACAGAATT-TTGAAA-3’; Antisense: 5’-AGCTTTTCCAAAAAATTCTGTGTGGTGTCACCTGTCTCTTGAACAGGTGACACCA-CACAGAAC-3’. The oligonucleotides for the insert were designed according to Ambion’s recommendations for short hairpin RNA (shRNA) design. Each insert contains a BamHI and a HindIII restriction site overhang, a 19 bp target sequence, a 9 bp loop sequence, and the reverse complementary sequence of the 19 bp target. The insert also includes an RNA polymerase III terminator sequence. The oligos for the insert were annealed at 1 μg/μl and 37°C for 1 h as specified in the kit.
A clone, pSCR, provided by Ambion, which expresses a shRNA scrambled sequence, was used as a control (Sense: 5’-GATCCACTACCGTTGTTAAGGTGTTCAAGAGACACCTATAACAACGGTAGTTTTTG GAAAA-3’). pSCR clones were transformed into competent DH5α E. coli cells and selected by growing in Luria Broth medium supplemented with 100 μg/ml of ampicillin. Plasmid DNA was isolated and purified using the Maxiprep kit (Qiagen) according to manufacturer’s instructions. The presence of plasmid constructs and insert sequences were confirmed by DNA sequencing.
Cell Culture and Maintenance
Human embryonic kidney (HEK 293) cells were obtained from American Type Culture Collection (ATCC) [cat #CRL1573] and cultured with DMEM supplemented with 10% heat-inactivated FBS, 1% 200 mM L-glutamine, and 1% sodium pyruvate. The cells were maintained in 100 mm tissue culture dishes and passaged 1:10 - 1:12 with trypsin/EDTA at 80% confluency. Human ovarian (SKOV-3) cells [ATCC; cat #HTB-77] were cultured with McCoy′s 5a medium supplemented with 10% FBS, 2 mM L-glutamine. Human mammary (MDA-MB-157) cancer cells [ATCC cat #HTB-24] were cultured with DMEM F12 50/50 medium supplemented with 10% FBS and 2 mM L-glutamine. All cultures were incubated at 37°C in 5% CO2.
Transfection and Selection; HEK cells
For dsRNA transfection, cells were trypsinized 24 h before transfection and viable cells were counted with a hemacytometer using the Trypan blue dye exclusion method. 4.0 × 105 cells were plated per well of a 6-well plate and incubated for 24 h to provide experimental cultures of approximately 60% confluency for transfection the next day. dsRNA (2.5 μg) was transfected in each well with FuGENE 6 transfection reagent [Roche, Indianapolis, IN] at a ratio of 1:5 μg to μl according to manufacturer’s recommendations. The FuGENE-dsRNA complex was allowed to incubate with the cells until harvesting and counted with a hemacytometer.
For plasmid DNA transfection, cells were transfected with pS2-3, and pSCR using FuGENE 6 transfection reagent [Roche] at a ratio of 1:5 μg to μl, according to manufacturer’s recommendations. The FuGENE-DNA complex was allowed to incubate with the cells for 24 h before removal and replenishing with fresh culture medium supplemented with antibiotics. Antibiotic resistant cells were selected with 300 μg/ml of G418 antibiotic in HEK medium. Cells were selected for at least 4 weeks before harvesting for analysis.
Transfection and Selection; SKOV-3 and MDA-MB-157 cells
Cells were transfected with pS2-3, and pSCR using FuGENE 6 transfection reagent [Roche] at a ratio of 1:3 μg to μl, according to manufacturer’s recommendations. The FuGENE-DNA complex was allowed to incubate with the cells for 24 h before removal and replenishing with fresh culture medium supplemented with antibiotics. Antibiotic resistant cells were selected with 800 μg/ml and 400 μg/ml of G418 antibiotic for SKOV-3 and MDA-MB-157 cells, respectively. Cells were selected for at least 4 weeks before harvesting for analysis.
Harvesting and Cell Counting
HEK cells transfected with dsRNA were harvested and counted at days 1, 3, and 5. HEK and SKOV-3 cells transfected with plasmid constructs were allowed to recover and selected for at least 4 weeks before harvesting for analysis and setting up of cell proliferation assays. Cell-proliferation assays were performed by trypsinizing the cells, using the Trypan blue dye exclusion method and counting with a hemacytometer.
RT-PCR Analysis
The expression of hTERT, p53, and p21 were assessed by PCR using cDNA libraries generated from treated cells. PCR analysis of transcripts for housekeeping genes, GAPDH and β-actin, were performed as loading controls and were used as standards to normalize data. Total RNA was extracted using the Qiagen RNeasy kit (Qiagen). Extracted RNA was quantified with a UV spectrophotometer and equal amounts of RNA per experiment were used to generate cDNA with the Superscript First-Strand Synthesis System (Invitrogen). Synthesized cDNA was subjected to PCR amplification with primers targeting the gene products for hTERT (F: 5’-CAAGAACGCAGGGATGTCGCTG-3’; R: 5’-CTGCGTCTGGGCTGTCCTGAGT-3’), p53 (F: 5’-ATTTGCGTGTGGAGTATTTG-3’; R: 5’-GGAACAAGAAGTGGAGAATG-3’), p21 (F: 5’-CAGGGTCGAAAACGGCGGCA-3’; R: 5’-AGGAGCCACACCCCTCCAGA-3’), GAPDH (F: 5’-GAAGGTGAAGGTCGGAGTC-3’; R: 5’-GAAGATGGTGATGGGATTTC-3’) and β-actin (F: 5’-GCTCGTCGTCGACAACGGCTC-3’; R: 5’-CAAACATGATCTGGGTCATCT TCTC-3’). The PCR reactions were analyzed by 2% agarose gel electrophoresis. For each primer set, the number of amplification cycles was predetermined to in order to be in the exponential phase.
Statistical Analysis
Cell counts were subjected to a two-tailed T-test, assuming equal variances. Mock transfected and transfected samples were compared to the control untransfected samples to determine significant differences (p<0.05) in proliferation rate.
Results
hTERT knock down using long dsRNA and shRNA
To assess the short-term effects of hTERT knock down, a 219 bp long dsRNA complementary to hTERT mRNA beginning at +863, was synthesized and transfected into HEK cells. hTERT mRNA in the cells was significantly reduced to 55% by 24 h of treatment and the RNAi slowly decreased in effectiveness thereafter (Fig. 1A). GAPDH was used as a loading control and its consistent expression levels show that the RNAi effect was specific for hTERT. To determine the effects of hTERT knock down on the cell-cycle, a growth curve was generated. Untreated HEK cells and cells incubated with only FuGENE 6 tranfection reagent were used as controls to ensure that there is no toxicity due to the transfection reagent. The proliferation studies showed parallel linear growth between the untreated and mock-treated control cells while treated cells were growth inhibited (Fig. 1B and C). Growth inhibition peaked on day 3 of treatment, and was sustained through day 6 after treatment (Fig. 1C).
Fig. 1.
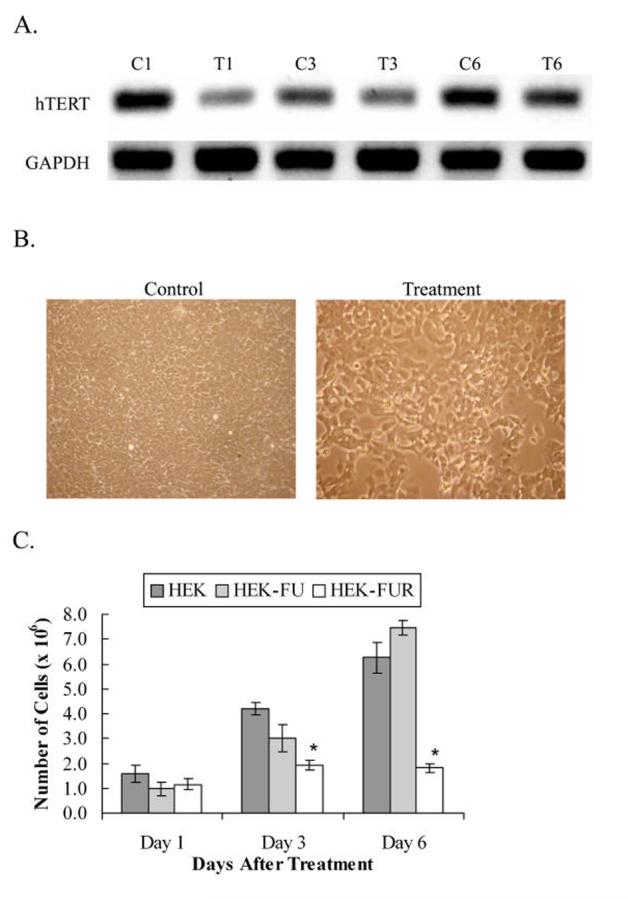
Knock down of hTERT in HEK cells from transfecting cells with dsRNA complementary to hTERT mRNA. A, PCR analysis of hTERT and GAPDH expression were performed using cDNA from treated and untreated HEK cells. C1, C3 and C6, correspond to HEK untreated control cells on treatment days 1, 3 and 6, respectively. T1, T3 and T6, correspond to HEK cells treated with 2.5 μg dsRNA complementary to a 219 bp section of hTERT mRNA on treatment days 1, 3 and 6, respectively. Levels of hTERT mRNA are reduced within 24 hours of treatment due to RNAi and the levels begin to slowly recover thereafter. GAPDH was used as a loading control. B, HEK control and dsRNA-treated HEK cells at a total magnification of 100X on day 6 after transfection. The panel with the control HEK cells shows a more densely populated culture than the treated cells. C, Cells plated at a density of 4 × 105 cells and counted on days 1, 3, and 6 after dsRNA transfection. HEK: untreated control cells. HEK-FU: control cells incubated with Fugene 6 transfection reagent. HEK-FUR: dsRNA-treated cells transfected using Fugene 6 transfection reagent. Y-axis error bars represent ± standard error of the mean (SEM). * denotes statistical significance (n = 3, p < 0.05).
The cellular mRNA levels of p53 and p21 were analyzed following the introduction of RNAi of hTERT on days 1, 3, and 6 post-treatment with the 219 bp dsRNA. p53 mRNA levels were elevated within 24 hours of treatment, correlating with the decreased levels of hTERT mRNA in the cell (Fig. 2A). p53 mRNA levels showed a marked 4-fold elevation in treated HEK cells on day 1 and remained elevated through day 6 (Fig. 2B), while the untreated HEK control cells showed a steady incline of p53 mRNA levels to 3.1 fold by day 6. This suggests that the increase in p53 levels in the control HEK cells probably resulted from cell-to-cell contact as opposed to a response to lowered hTERT levels. Following the induction of p53 in the treated HEK cells, p21 mRNA levels progressively increased, consistent with findings that p53 transactivates the p21 promoter [23, 24] (Fig. 2A). In the treated HEK cells, p21 mRNA levels were increased 1.7 fold on day 1 and 4.6 fold by day 6 (Fig. 2C). In contrast, p21 levels in the untreated HEK control cells were slightly elevated to 1.5 fold and remained relatively stable (Fig. 2C). Further, since HEK cells do not elicit an interferon response when transfected with long dsRNA [27, 28] and the GAPDH loading control does not show a variable expression, the results indicate that the RNAi response is target specific and appropriated to the dsRNA treatment, instead of off-target RNAi effects. Taken together, the findings suggest that reduced levels of hTERT mRNA in HEK cells resulted in a profound growth inhibitory response induced by the p53/p21 pathway.
Fig. 2.
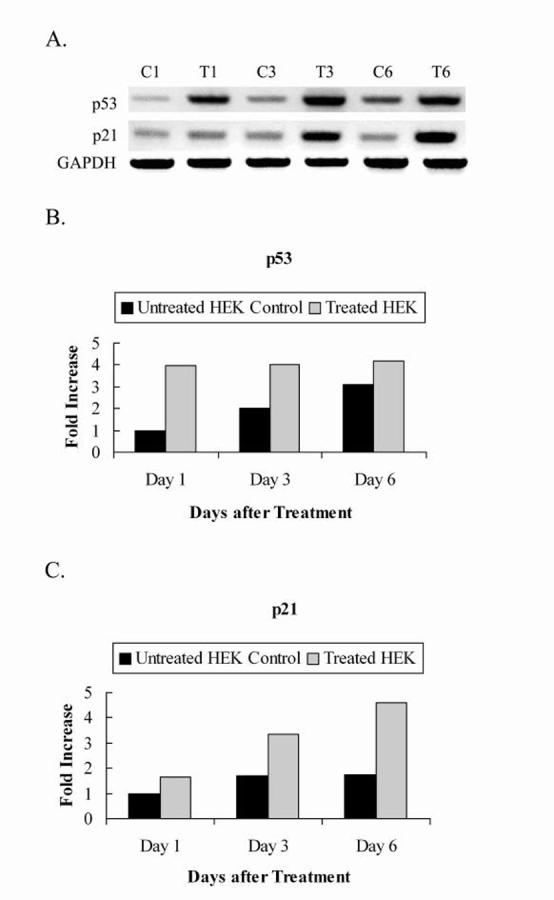
PCR analysis of p53, p21 and GAPDH expression using cDNA from HEK cells treated and untreated with synthesized dsRNA. A, 2% agarose gel electrophoresis showing p53 and p21 mRNA levels after induction of RNAi of hTERT. GAPDH was included as a loading control. Lanes C1, C3, and C6: untreated HEK control cells at days 1, 3, and 6 after treatment respectively. Lanes T1, T3, and T6: treated HEK cells at days 1, 3, and 6 after treatment respectively. B, C, Quantitations of p53 and p21 mRNA levels respectively, normalized to housekeeping gene, GAPDH. Values are from a representative gel.
The plasmid construct, pS2-3, was transfected into HEK cells and selected in G418 antibiotic for at least 4 weeks to analyze the long-term effects of hTERT knock down. The experiment was repeated twice to ensure reproducibility of results. Non-transfected HEK cells and HEK cells transfected with a construct expressing a scrambled sequence (pSCR) were used as controls. Proliferation studies showed comparative linear growth in both the control HEK and pSCR cells. HEK cells expressing pS2-3 showed slower growth rates as compared to the controls (Fig. 3E). To determine whether the constructs had successfully knocked-down hTERT and the effects on other genes of interest, total RNA was extracted from untransfected HEK cells and HEK transfected with pS2-3. cDNA was synthesized via reverse transcriptase reactions and PCR was performed with primers specific to hTERT, p53, p21, GAPDH, and β-actin. Cells transfected with pS2-3 showed decreased levels of hTERT mRNA as compared to the control cells, indicating that the construct was able to, but not fully knock-down hTERT (Fig. 3A).
Fig. 3.
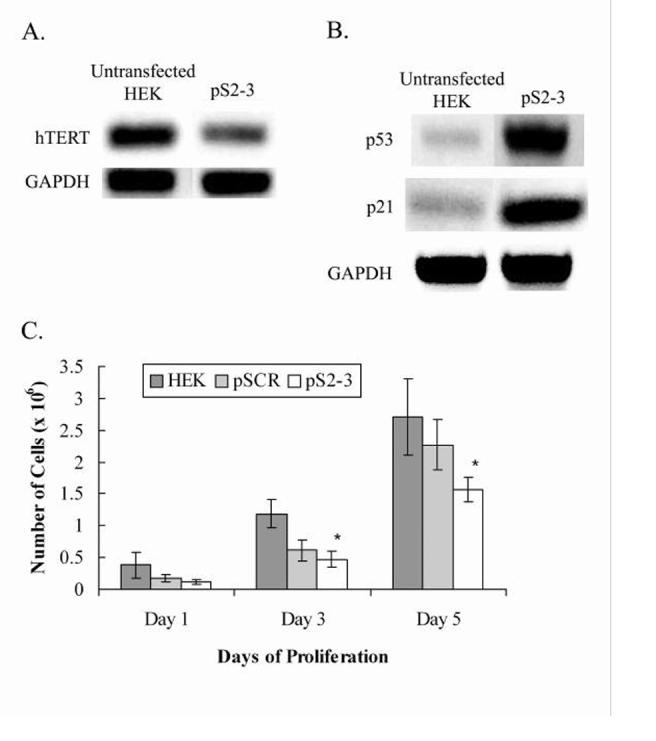
Gene expression and proliferation assays of control HEK cells and HEK cells transfected with plasmid constructs pSCR and pS2-3. A, hTERT mRNA levels showing hTERT knock down in cells transfected with pS2-3 as compared to the untransfected HEK. GAPDH was used as a loading control. B, Increased transcription of p53 and p21 in cells transfected with pS2-3 as compared to the untransfected HEK cells. GAPDH was used as a loading control. C, The graph shows linear growth of controls HEK and pSCR, while pS2-3 shows a significantly slower growth rate. Y-axis error bars represent ± standard error of the mean (SEM). * denotes statistical significance (n = 3, p < 0.05).
The effect of hTERT knock down on the other genes of interest paralleled that of the short-term studies with dsRNA transfection. As expected, HEK cells transfected with pS2-3 showed elevated levels of p53 and p21 (Fig. 3C). Further, the invariability in expression of the housekeeping gene, GAPDH, indicates that the knock down is target specific. Although hTERT was not fully knocked-down, it is important to note that it is adequate to affect changes on the expression of p53 and p21. Taken together, the results of long-term knock down of hTERT in HEK cells suggest that telomerase may directly or indirectly affect the cell cycle or the genes regulating the cell cycle.
hTERT knock down in p53 null SKOV-3 and MDA-MB-157 cells
To investigate the knock down of hTERT in p53-null systems in order to eliminate the variable of p53-induced effects on p21, pS2-3 and pSCR plasmid constructs were transfected into SKOV-3 ovarian cancer cells and MDA-MB-157 breast cancer cells. Cells transfected with plasmid constructs were subjected to G418 antibiotic selection for at least a month before harvesting for analysis.
Untransfected SKOV-3 cells (denoted as SKOV) and SKOV-3 cells transfected with the plasmid expressing a scrambled sequence (SKOVpSCR) were used as controls. However, in our proliferation studies, SKOVpS2-3 cells did not show a marked retardation in growth, as compared to SKOV and SKOVpSCR. By day 3 of the proliferation studies, SKOV, SKOVpSCR, and SKOVpS2-3 cells seemed to reach a plateau in the rate of proliferation after becoming confluent and experiencing contact inhibition in culture. There was no obvious growth in these cultures between day 3 and day 5 (Fig. 4B). This is consistent with a previous study that showed that inhibition of telomerase activity in SKOV-3 cells neither resulted in telomeric erosion nor growth inhibition [36].
Fig. 4.
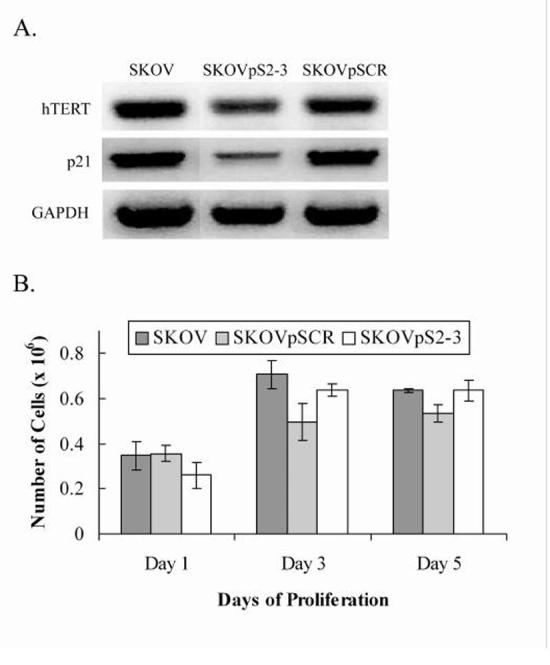
Gene expression and proliferation assays of SKOV, SKOVpS2-3, and SKOVpSCR. A, Reverse transcriptase PCR of RNA extracts from untransfected SKOV, SKOVpS2-3, and SKOVpSCR. SKOV and SKOVpSCR were used as controls. PCR was performed with primers specific to the gene of interest: hTERT, p21, and GAPDH. GAPDH was used as a loading control. B, No significant inhibition of growth was observed in association with hTERT knock down in SKOVpS2-3 (n = 3, p < 0.05).
To determine if hTERT had been successfully knocked-down in the transfected SKOV-3 cells, reverse transcriptase PCR analysis was performed and showed hTERT knock down in SKOVpS2-3 (Fig. 4A). Corresponding to decreased levels of hTERT mRNA in SKOVpS2-3 cells, expression of p21 also appeared to be down-regulated (Fig. 4A), while p21 expression was not affected in the controls SKOV and SKOVpSCR. The invariability of the expression of GAPDH also indicates that the effect of RNAi is target specific.
MDA-MB-157 cells transfected with pS2-3 (MDA-pS2-3) showed a significant reduction in proliferation rate as compared to control untransfected MDA-MB-157 cells and cells expressing the scrambled sequence construct (MDA-pSCR) (Fig. 5C). This indicates that hTERT protein or hTERT mRNA is able to exert control on cell cycle progression either directly or indirectly.
Fig. 5.
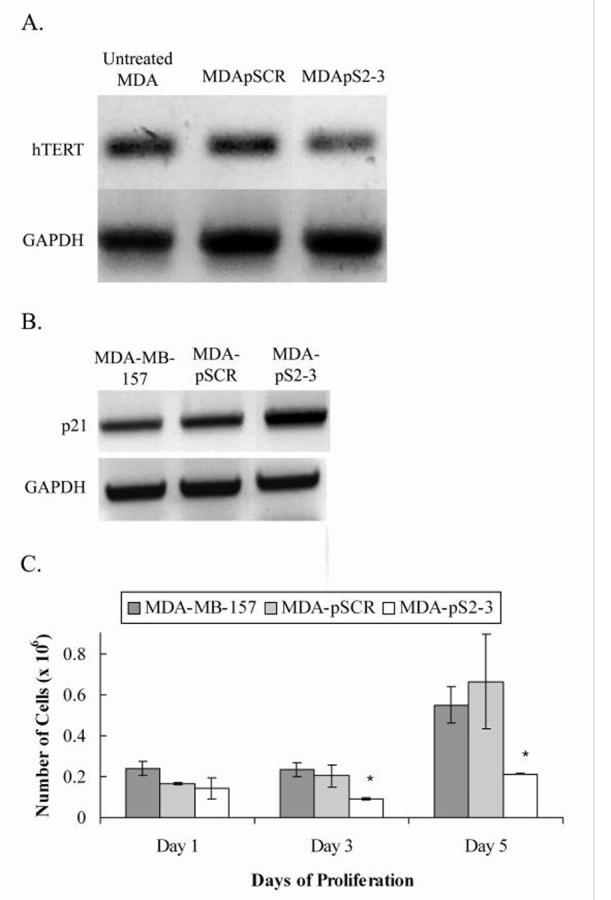
Gene expression and proliferation assays of MDA-MB-157 untreated control cells and cells transfected with plasmid constructs pSCR and pS2-3 (MDA-pSCR and MDA-pS2-3 respectively). A, Reverse transcriptase PCR of RNA extracts from treated MDA-MB-157 cells and untreated cells at 74 days after transfection. PCR was performed with primers specific to the gene of interest. hTERT mRNA levels showing hTERT knock down in MDA-pS2-3 as compared to the untransfected MDA-MB-157 and MDA-pSCR control cells. GAPDH was used as a loading control. B, Increased transcription of p21 was observed in pS2-3 as compared to the untransfected controls MDA-MB-157 and MDA-pSCR. GAPDH was used as a loading control. C, The graph shows parallel growth of controls MDA-MB-157 and MDA-pSCR, while MDA-pS2-3 shows growth inhibition. Y-axis error bars represent ± standard error of the mean (SEM). * denotes statistical significance (n = 3, p < 0.05).
Reverse transcriptase PCR using primers specific to the genes of interest was performed to determine effect of hTERT knock down on the levels of transcription for hTERT and p21. The knock down was confirmed by lower levels of hTERT transcript in MDA-pS2-3 cells but not in control MDA-MD-157 or MDA-pSCR cells (Fig. 5A). The downstream effects of the knock down resulted in an increase in p21 expression in MDA-pS2-3 while expression of p21 in control cells MDA-MB-157 and MDA-pSCR remained at basal levels (Fig. 5B).
Discussion
Telomerase has only recently been found to exert extra-telomeric effects [17]. Here, we show specific effects of hTERT expression and telomerase activity on the cell cycle regulators, p53 and p21. Both of these genes are known upstream regulators of hTERT, suggesting the concept that one of the extra-telomeric effects of hTERT may be in a feedback loop, thereby regulating itself. As there are multiple regulatory factors that act on the hTERT promoter, such as epigenetic modifications, p53, p21, c-Myc/Mad/Max, estrogen-receptor complexes, Sp1, WT1, E2F [37-42], it is the culmination of all factors exerting their effects on the hTERT promoter that eventually determines the transcriptional output of hTERT. Several studies have also successfully knocked down hTERT and shown that RNAi of hTERT decreases telomerase activity, indicating that RNAi if hTERT is a viable system to study the downstream effects of telomerase [43-45]. Our results suggest that the effects of hTERT and telomerase activity on extra-telomeric targets vary in different cellular systems.
Since HEK cells are of embryonic origin, we were able to use two different RNAi systems to knock down hTERT. Namely, dsRNA and a plasmid construct expressing an shRNA, were used to knockdown hTERT mRNA. Since both RNAi systems in HEK cells generated similar results, we are able to conclude that the HEK cells did not experience interferon response due to the transfection of long dsRNA, and that the RNAi effects were target specific. Moreover, in all experiments, we observed no variability in expression of housekeeping genes, GAPDH and β-actin, further supporting that the RNAi was target-specific.
The knock down of hTERT using dsRNA targeting a 219 bp region of the hTERT mRNA resulted in a significant reduction of hTERT mRNA in the cell, and subsequently caused the cells to be temporarily growth arrested. The RNAi effect on hTERT caused an immediate increase in p53 mRNA levels, which in turn induced the transcription of p21 [23, 24] (Table 1), suggesting that some specific telomeric-effects of hTERT include the regulation of p53 and p21. It is also highly unlikely that changes in telomeric lengths alone affected cell proliferation rate and induced the upstream regulators of hTERT. Instead, it is probable that the knock down of hTERT itself induced such effects, because p53 and p21 levels were elevated in the transient dsRNA knock down of hTERT within 24 h of treatment, before telomeric changes are detectable. Thus, telomerase is able to exert an effect on the cell cycle independent of telomere length.
Table 1.
Summary of proliferation changes and expression of p53 and p21 in cell lines HEK, SKOV, and MDA-MB-157.
| Cell Line | Genotype | Effects of hTERT knock down |
||
|---|---|---|---|---|
| Proliferation | p53 | p21 | ||
| HEK | Adenoviral transformed | ↓ | ↑ | ↑ |
| MDA-MB-157 | p53-/- | ↓ | - | ↑ |
| SKOV-3 | p53-/-; p16INK4a-/- | - | - | ↓ |
When 19-mer shRNAs targeting hTERT mRNA were used to stably knock down hTERT in long-term studies, we observed differential responses to the RNAi effect. In HEK cells, the pS2-3 construct expressing shRNA specific to hTERT retarded growth of the cells and caused an elevation of p53 and p21 mRNA in response to hTERT knock down. The finding that the cells were not growth-arrested could be attributed to the fact that the RNAi knock down effect using a single short target was not as effective as targeting hTERT with the 219 bp dsRNA. Also, the residual hTERT levels after knock down may provide enough telomerase activity to sustain proliferation, despite its slowed rate. It is also significant to note that although the knock down of hTERT was not 100%, the slight reduction in hTERT mRNA levels was adequate to produce changes in proliferation rate and exert extra-telomeric effects on p53 and p21.
To further evaluate how hTERT might affect p53-null and different neoplastic cell systems, hTERT was knocked down in SKOV-3 ovarian cancer and MDA-MB-157 breast cancer cells. Proliferation studies showed a significant growth decrease in MDA-pS2-3 cells expressing shRNA specific to hTERT mRNA as compared to the untreated control cells as well as the cells expressing a scrambled shRNA sequence (MDA-pSCR). This cell growth reduction was coupled with an increase in p21 mRNA levels, consistent with the HEK study. It is very probable that the growth reduction is due to the upregulation of p21. This also indicates that hTERT is able to regulate p21 independent of p53.
SKOV-pS2-3 cells expressing shRNA specific to hTERT mRNA resulted in a decreased expression of p21, supporting the evidence that hTERT can regulate p21 in a p53-independent manner. It is interesting that we did not observe a significant retardation of growth in SKOVpS2-3 cells as compared to the untreated control cells and the cells expressing a scrambled shRNA sequence (SKOVpSCR). Further, since SKOV-3 cells do not undergo alternate lengthening of telomeres (ALT)[36], a probable explanation for the lack of growth reduction could be that since SKOV-3 cells are null for both p53 and p16INK4a, a decrease in p21 due to the knock down may result in insufficient cell cycle inhibitory machinery. However, the exact mechanisms of how hTERT up or down regulates p21 in these systems remain unclear.
Our findings support the novel model that telomerase, via the regulation of hTERT (the gene encoding its catalytic subunit), is involved in a feedback loop that may engage various pathways. Our studies clearly indicate that the knock down of hTERT is able to affect its own regulatory gene components, namely, p53 and p21. In cell systems such as HEK cells where the p53 gene is intact, the feedback loop is p53 dependent resulting in p21 transcriptional activation and cell growth inhibition. In p53-null cell systems such MDA-MB-157 cells, the knock down of hTERT results in a p53-independent upregulation of p21, with similar growth retardation. In contrast, the knock down of hTERT in p53/p16INK4a-null and ALT-negative SKOV-3 cells resulted in a decreased expression of p21 and did not result in growth inhibition.
These findings indicate that perhaps the titer of hTERT mRNA or telomerase activity in the cell may signal the cell to induce a feedback regulation system, also further supporting the hypothesis that telomerase has extra-telomeric functions. The precise mechanisms of the feedback system need to be refined and further detailed. Future studies involving other cell types and the use of a p21-null model would be useful to clarify how other genes are affected after hTERT knock down.
Acknowledgements
We gratefully acknowledge Dr. Lynn Dobrunz for kindly offering the use of laboratory equipment and resources, and Dr. Robert Angus for his expert statistical consultation. We also kindly thank Brandon Walters and James Dekay for their expert assistance and support during the project. This work was funded in part by a grant from the NCI, a grant from the Susan G. Komen Breast Cancer Foundation, an NIH Ovarian SPORE grant, and an Evelyn F. McKnight Brain Institute grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Nakayama J, Tahara H, Tahara E, Saito M, Ito K, Nakamura H, Nakanishi T, Ide T, Ishikawa F. Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nat. Genet. 1998;18:65–68. doi: 10.1038/ng0198-65. [DOI] [PubMed] [Google Scholar]
- [2].Armelin HA, Armelin MCS, Kelly K, Stewart T, Leder P, Cochran BH, Stiles CD. Functional role for c-myc in mitogenic response to platelet-derived growth factor. Nature (London) 1984;310:655–660. doi: 10.1038/310655a0. [DOI] [PubMed] [Google Scholar]
- [3].Vaziri H, Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr. Biol. 1998;8:279–282. doi: 10.1016/s0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- [4].Wright WE, Shay JW. Two-step mechanism controlling cellular senescence and immortalization. Exp. Geront. 1992;27:383–389. doi: 10.1016/0531-5565(92)90069-c. [DOI] [PubMed] [Google Scholar]
- [5].Greider CW. Telomeres. Curr. Opin. Cell Biol. 1991;3:444–451. doi: 10.1016/0955-0674(91)90072-7. [DOI] [PubMed] [Google Scholar]
- [6].Greider CW, Blackburn EH. Telomeres, telomerase and cancer. Sci. Am. 1996;274:92–97. doi: 10.1038/scientificamerican0296-92. [DOI] [PubMed] [Google Scholar]
- [7].Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- [8].Greider CW, Blackburn EH. The telomere terminal transferase of tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- [9].Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- [10].Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Baijersbergen RL, Davidoff MJ, Liu Q, Bacchetti S, Haber DA, Weinberg RA. hEST2, the putative human telomerase catalytic subunit gene, is upregulated in tumor cells during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- [11].Nakamura TN, Morin G, Chapman KB, Wienrich RL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and humans. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- [12].Matsutomi K, Yu EY, Khurts S, Ben-Porath I, Currier JL, Metz GB, Brooks MW, Kaneko S, Murakami S, DeCaprio JA, Weinberg RA, Stewart SA, Hahn WC. Telomerase maintains telomere structure in normal human cells. Cell. 2003;114:241–253. doi: 10.1016/s0092-8674(03)00550-6. [DOI] [PubMed] [Google Scholar]
- [13].Yi X, Tesmer VM, Savre-Train I, Shay JW, Wright WE. Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol. Cell. Biol. 1999;19:3989–3997. doi: 10.1128/mcb.19.6.3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li M, Wu Y, Liang YR, Wu XY. Correlation between expression of human telomerase subunits and telomerase activity in esophageal squamous cell carcinoma. World J. Gastroenterol. 2003;9:2395–2399. doi: 10.3748/wjg.v9.i11.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cong Y-S, Wen J, Bacchetti S. Human telomerase and its regulation. Microbiol. Molec. Biol. Rev. 1999;66:407–425. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ducrest AL, Szutorisz H, Lingner J, Nabholz M. Regulation of the human telomerase gene. Oncogene. 2002;21:541–552. doi: 10.1038/sj.onc.1205081. [DOI] [PubMed] [Google Scholar]
- [17].Perrault SD, Hornsby PJ, Betts DH. Global gene expression response to telomerase in bovine adrenocortical cells. Biochem. Biophys. Res. Commun. 2005;335:925–936. doi: 10.1016/j.bbrc.2005.07.156. [DOI] [PubMed] [Google Scholar]
- [18].Kanaya T, Kyo S, Hamada K, Takakura M, Kitagawa Y, Harada H, Inoue M. Adenoviral expression of p53 represses telomerase activity through down-regulation of human telomerase reverse transcriptase transcription. Clin. Cancer Res. 2000;6:1239–1247. [PubMed] [Google Scholar]
- [19].Dotto GP. More than a break to the cell cycle? Biochim. Biophys. Acta. 2000;1471:M43–M56. doi: 10.1016/s0304-419x(00)00019-6. [DOI] [PubMed] [Google Scholar]
- [20].Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:634–635. [Google Scholar]
- [21].Harada K, Kurisu K, Sadamoto T, Tahara H, Tahara E, Ide T, Tahara E. Growth inhibition of human glioma cells by transfection-induced p21 and its effects on telomerase activity. J. Neurooncol. 2000;47:39–46. doi: 10.1023/a:1006428529637. [DOI] [PubMed] [Google Scholar]
- [22].Hendersen YC, Breau RL, Liu TJ, Clayman GL. Telomerase activity in head and neck tumors after introduction of wild-type p53, p21, p16, E2F-1 genes by means of recombinant adenovirus. Head Neck. 2000;22:347–354. doi: 10.1002/1097-0347(200007)22:4<347::aid-hed6>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- [23].Koutsodontis G, Tentes I, Papakosta P, Moustakas A, Kardassis D. Sp1 plays a critical role in the transcriptional activtation of the human cyclin-dependent kinase inhibitor p21WAF1/Cip1 gene by the p53 tumor suppressor protein. J. Biol. Chem. 2001;276:29116–29125. doi: 10.1074/jbc.M104130200. [DOI] [PubMed] [Google Scholar]
- [24].Prowse DM, Bolgan L, Molnár A, Dotto GP. Involvement of the Sp3 transcription factor in induction of p21Cip1/WAF1 in keratinocyte differentiation. J. Biol. Chem. 1997;272:1308–1314. doi: 10.1074/jbc.272.2.1308. [DOI] [PubMed] [Google Scholar]
- [25].Young JI, Smith JR. DNA methyltransferase inhibition in normal human fibroblasts induces a p21-dependent cell cycle withdrawal. J. Biol. Chem. 2001;276:19610–19616. doi: 10.1074/jbc.M009470200. [DOI] [PubMed] [Google Scholar]
- [26].Louis N, Evelegh C, Graham FL. Cloning and sequencing of cellular viral junctions from the human adenovirus type 5 transformed 293 cell line. Virology. 1997;233:423–429. doi: 10.1006/viro.1997.8597. [DOI] [PubMed] [Google Scholar]
- [27].Ali S, Kukolj G. Interferon regulatory factor 3-independent double-stranded RNA-induced inhibition of hepatitis C virus replicons in human embryonic kidney 293 cells. J. Virology. 2005;79:3174–3198. doi: 10.1128/JVI.79.5.3174-3178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yang S, Tutton S, Pierce E, Yoon K. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol. Cell. Biol. 2001;21:7807–7816. doi: 10.1128/MCB.21.22.7807-7816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Billy E, Brondani V, Zhang H, Müller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. PNAS. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yew PR, Berk AJ. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- [31].Chen J, Lin J, Levine AJ. Regulation of transcription functions of the p53 tumor suppressor by the mdm-2 oncogene. Mol. Med. 1995;1:142–152. [PMC free article] [PubMed] [Google Scholar]
- [32].Chen J, Wu X, Levine AJ. Mdm-2 inhibits G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol. Cell. Biol. 1996;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Löber C, Lenz-Stöppler C, Dobbelstein M. Adenovirus E1-transformed cells grow despite the continuous presence of transcriptonally active p53. J. Gen. Virol. 2002;83:2047–2057. doi: 10.1099/0022-1317-83-8-2047. [DOI] [PubMed] [Google Scholar]
- [34].Wick M, Zubov D, Hagen G. Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT) Genes Dev. 1999;232:97–106. doi: 10.1016/s0378-1119(99)00108-0. [DOI] [PubMed] [Google Scholar]
- [35].Ramirez PT, Gershenson DM, Guillermo T-L, Ramondetta LM, Fightmaster D, Wharton JT, Wolf JK. Expression of cell-cycle mediators in ovarian cancer cells after transfection with p16INK4a, p21WAF1/Cip-1, and p53. Gyn. Onc. 2001;83:543–548. doi: 10.1006/gyno.2001.6438. [DOI] [PubMed] [Google Scholar]
- [36].Gan Y, Mo Y, Johnston J, Lu J, Wientjes MG, Au JL-S. Telomere maintenance in telomerase-positive human ovarian SKOV-3 cells cannot be retarded by complete inhibition of telomerase. FEBS Lett. 2002;527:10–14. doi: 10.1016/s0014-5793(02)03141-1. [DOI] [PubMed] [Google Scholar]
- [37].Shats I, Milyavsky M, Tang X, Stambolsky P, Erez N, Brosh R, Kogan I, Braunstein I, Tzukerman M, Ginsberg D, Rotter V. p53-dependent down-regulation of telomerase is mediated by p21waf1. J. Biol. Chem. 2004;279:50976–50985. doi: 10.1074/jbc.M402502200. [DOI] [PubMed] [Google Scholar]
- [38].Xiang H, Wang J, Mao Y, Liu M, Reddy VN, Li DW. Human telomerase accelerates growth of lens epithelial cells through regulation of the genes mediating RB/E2F pathway. Oncogene. 2002;21:3784–3791. doi: 10.1038/sj.onc.1205455. [DOI] [PubMed] [Google Scholar]
- [39].Dean M, Levine RA, Campisi J. c-myc regulation during retinoic acid-induced differentiation of F9 cells is posttranscriptional and associated wtih growth arrest. Mol. Cell. Biol. 1986;6:518–524. doi: 10.1128/mcb.6.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Won J, Yim J, Kim TK. Opposing regulatory roles of E2F in human telomerase reverse transcriptas (hTERT) gene expression in human tumor and normal somatic cells. FASEB J. 2002;16:1943–1945. doi: 10.1096/fj.02-0311fje. [DOI] [PubMed] [Google Scholar]
- [41].Liu L, Lai S, Andrews LG, Tollefsbol TO. Genetic and epigenetic modulation of telomerase activity in development and disease. Genes Dev. 2004;340:1–10. doi: 10.1016/j.gene.2004.06.011. [DOI] [PubMed] [Google Scholar]
- [42].Poole JC, Andrews LG, Tollefsbol TO. Activity, function, and gene regulation of the catalytic subunit of telomerase (hTERT) Gene. 2001;269:1–12. doi: 10.1016/s0378-1119(01)00440-1. [DOI] [PubMed] [Google Scholar]
- [43].Li Y, Li M, Peng Y, Jiang Z, Li W, Li H. Suppression of telomerase acitivty by plasmid-mediated RNA interference. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2006;23:615–619. [PubMed] [Google Scholar]
- [44].Zheng JN, Sun YF, Pei DS, Liu JJ, Li CJC,W, Sun XQ, Shi QD, Han RF, Ma TX. Inhibition of proliferation and induction of apoptosis in human renal carcinoma cells by anti-telomerase small interfering RNAs. Acta Biochim Biophys Sin (Shanghai) 2006;38:500–506. doi: 10.1111/j.1745-7270.2006.00182.x. [DOI] [PubMed] [Google Scholar]
- [45].de Souza Nascimento P, Alves G, Fiedler W. Telomerase inihibition by an siRNA directed against hTERT leads to telomere attrition in HT29 cells. Oncol. Rep. 2006;16:423–428. [PubMed] [Google Scholar]


