Abstract
Hematopoietic stem cells (HSCs) execute self-renewal divisions throughout fetal and adult life, although some of their properties do alter. Here we analyzed the magnitude and timing of changes in the self-renewal properties and differentiated cell outputs of transplanted HSCs obtained from different sources during development. We also assessed the expression of several “stem cell” genes in corresponding populations of highly purified HSCs. Fetal and adult HSCs displayed marked differences in their self-renewal, differentiated cell output, and gene expression properties, with persistence of a fetal phenotype until 3 weeks after birth. Then, 1 week later, the HSCs became functionally indistinguishable from adult HSCs. The same schedule of changes in HSC properties occurred when HSCs from fetal or 3-week-old donors were transplanted into adult recipients. These findings point to the existence of a previously unrecognized, intrinsically regulated master switch that effects a developmental change in key HSC properties.
Keywords: bone marrow transplantation, development, gene expression, self-renewal
The existence of hematopoietic stem cells (HSCs) was first demonstrated in the 1950s in experiments that revealed the ability of bone marrow cells from adult mice to permanently regenerate the blood-forming system of transplanted irradiated recipients (1). Later, clonal analysis of the regenerated progeny of uniquely tagged cells (2, 3) and, more recently, analysis of highly purified single-cell transplants (4, 5) have established the long-term self-renewal ability of a rare subset of hematopoietic cells in adult bone marrow that have unrestricted lymphomyeloid differentiation potential. Endpoints that detect the output of mature B cell, T cell, and myeloid blood cells in transplanted mice for at least 4 months allow these HSCs to be specifically identified in uncharacterized suspensions. Coupling such endpoints to limiting dilution transplantation protocols provides a powerful method for quantifying HSCs as competitive repopulating units (CRUs) (6).
HSCs detectable as CRUs are present throughout life and are believed to be generated by continuous self-renewal divisions from the first HSCs that, in mice, appear on embryonic day 9 (E9) (7, 8). Nevertheless, many elements of HSC behavior and the specific differentiation programs used by their maturing progeny change during the course of development (9–14). For example, transplants of cells from fetal liver regenerate daughter HSCs in irradiated recipients more quickly and outcompete the production of HSCs from adult bone marrow cells (10, 11). They also change their relative outputs of lymphoid and myeloid cells between fetal life and during the process of aging (13, 14). Recently, we showed that all HSCs in mice are actively cycling until 3 weeks after birth, at which time they abruptly switch, within 1 week, to become a predominantly quiescent population (15). These latter findings likely explain the reported differences in several phenotypic properties of HSCs that change during ontogeny; e.g., expression of CD34, CD38, and Mac1, and an ability to efflux Rhodamine-123 (Rho) and Hoechst 33342 (reviewed in ref. 16). These observations prompted us to design a series of experiments to compare the self-renewal activity, lineage biases, and gene expression profiles of HSCs isolated from tissues at various stages of development from E14.5 in fetal life to 12 weeks after birth. The results demonstrate that these properties are all dramatically and coordinately altered between 3 and 4 weeks after birth, indicating the activation of a developmental checkpoint that reprograms multiple key HSC functions at that time.
Results and Discussion
Quantitative Assessment of the Different HSC Regenerative Activity of Transplanted Cells Taken from Different Stages of Development.
To quantify the differences in the self-regenerative activity of fetal and adult HSCs, we compared their expansion kinetics in transplanted irradiated hosts by performing quantitative limiting dilution transplantation assays (CRU assays) of the regenerated cells in secondary irradiated hosts (see Fig. 1A). The results (Fig. 1B) confirmed the expected faster rate of expansion of the transplanted fetal liver HSCs as compared with transplants of adult bone marrow HSCs (17). We saw the greatest difference in HSC expansion rates during the second week posttransplant (when the number of fetal liver-derived HSCs increased 8-fold, whereas the number of adult marrow-derived HSCs remained largely unchanged). Moreover, in mice transplanted with equal numbers of genetically distinguishable HSCs from both sources, we found the rate of expansion of HSC numbers from each to be unaffected by cotransplanted cells from the other source (data not shown). Thus, the unique regeneration kinetics displayed by fetal and adult HSCs likely reflect intrinsically determined differences in their properties.
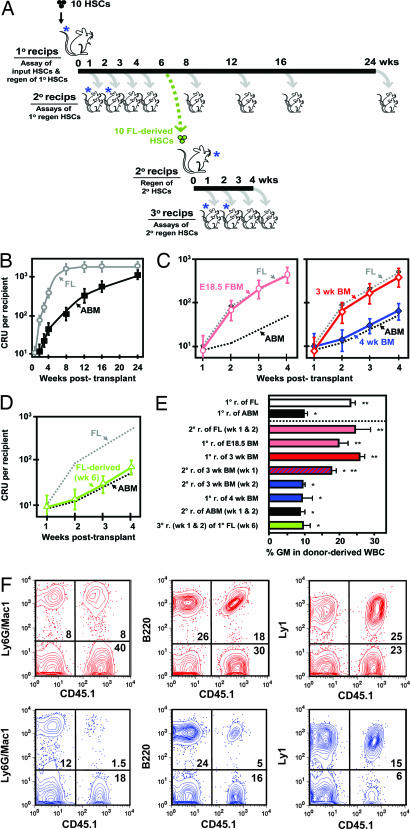
Fig. 1.
Rates of expansion and differentiation properties in vivo of HSCs reflect their developmental status. (A) Experimental design used to assess HSC regeneration kinetics in serially transplanted mice. In each HSC regeneration experiment, the initial transplant of unseparated cells contained ≈10 HSCs based on prior determination of the HSC content of the test cell suspension being used. These cells were then injected into sublethally irradiated Ly5-congenic W41/W41 recipients. This experimental design was used to maximize the contribution of the transplanted HSCs to the total HSC pool being regenerated while still allowing donor HSCs to be definitively identified, and also to remove any influences related to variable input HSCs doses that have been previously reported (17). The blue asterisks denote the groups of mice whose cells were analyzed 16 weeks posttransplant to demonstrate changes in the proportional contribution of mature myeloid cells to the total WBCs generated from the transplanted HSCs. (B) Comparison of HSC regeneration (mean ± SEM CRUs/mouse, four experiments each) from E14.5 fetal liver (FL, gray symbols) and adult bone marrow (ABM, black symbols) in sublethally irradiated W41/W41 mice injected initially with 10 HSCs (CRUs). (C) Similar analyses of mice transplanted with 10 HSCs (CRUs) from E18.5 fetal bone marrow (E18.5 FBM, pink symbols) and 3- and 4-week postnatal bone marrow (3-wk BM, red symbols; 4-wk BM, blue symbols). (D) HSCs (CRUs) produced from the HSC progeny of E14.5 fetal liver HSCs after 6 weeks in primary recipients (FL-derived, green symbols). (E) Relative outputs of donor-derived lymphoid and myeloid WBCs 16 weeks posttransplant in recipients of 3–6 CRUs. Each bar shows the mean ± SEM percentage of all donor-derived WBCs that expressed Ly6g and/or Mac1 (GM, n = 4–24). The information in the brackets in the labels for each bar refers to the number of weeks that were allowed to elapse before cells were harvested from primary recipients injected with the initial cells indicated and then transferred to secondary (or, in the case of the bottom bar, also into tertiary) recipients where the final lineage output assessments were made 16 weeks later. ∗, Values significantly lower (P < 0.05) than the value obtained for transplants of E14.5 fetal liver HSCs (CRUs). ∗∗, Values significantly higher (P < 0.05) than the value obtained for transplants of adult bone marrow HSCs (CRUs). (F) Representative FACS plots of the WBCs in mice 16 weeks after being transplanted with HSCs from 3-week (red) or 4-week (blue) postnatal bone marrow.
To investigate the timing and pace of the change in HSC self-regenerative activity during ontogeny, we repeated the experiment shown in Fig. 1B by using equivalent initial transplants of bone marrow cells from either E18.5 fetal mice or mice killed 3 or 4 weeks after birth. We then quantified the kinetics of donor-derived HSC regeneration in the bone marrow of the primary recipients of these cells as before by limiting dilution CRU assays of the cells in secondary mice. These measurements showed that HSCs from the bone marrow of E18.5 fetal and 3-week postnatal mice expanded with the same kinetics as HSCs from E14.5 fetal liver. In contrast, HSCs from the bone marrow of 4-week postnatal mice behaved similarly to the HSCs from the marrow of adult (8- to 12-week-old) mice (Fig. 1C). We also observed a marked change in the regenerative properties of HSCs derived from E14.5 fetal liver HSCs that had been transplanted directly into primary adult recipients 6 weeks previously. This result was obtained when the cells regenerated in the primary mice were transplanted into secondary mice and the rate of expansion of their progeny HSCs determined by performing CRU assays in tertiary recipients (Fig. 1D). Thus, within the same time frame (4–6 weeks), the progeny of E14.5 fetal liver HSCs acquire the regenerative properties of adult HSCs regardless of the age or physiological conditions of the host environment in which they are being produced.
Fetal and Adult HSCs Have the Same Cell Cycle Transit Times.
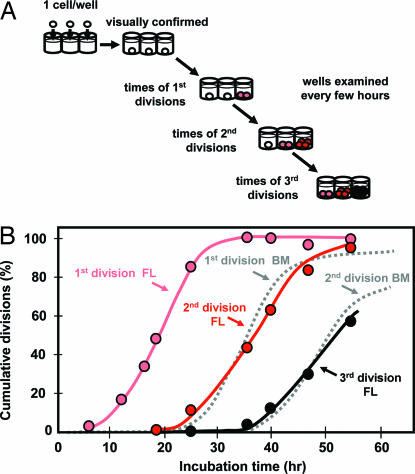
Differences in the rates of fetal and adult HSC expansion in irradiated recipients could reflect differences in the control of asymmetric versus symmetric HSC divisions, differences in HSC cell cycle transit times, and/or differences in the regulation of HSC viability. Evidence arguing against the latter two possibilities was obtained from experiments in which we tracked highly purified E14.5 fetal liver HSCs [see supporting information (SI) Fig. 5 A and D] in single-cell cultures under conditions that maintain fetal HSC numbers for 7 days (18). Under these conditions, we found that none of the input cells died, and they divided with the same average 14-hour cell cycle transit time (Fig. 2) as previously demonstrated for adult bone marrow HSCs, but without the significant delay the latter show due to their initial quiescent status (5). Interestingly, these studies also showed that the timing of the second and third divisions of fetal HSCs was the same as the timing of the first and second divisions, respectively, of adult bone marrow HSCs. Thus, fetal and adult HSCs cultured under conditions that have similar effects on their survival and self-renewal share the same cell cycle transit time once they have entered the cell cycle. Assuming similar conditions of HSC stimulation are operative in irradiated mice, a higher frequency of symmetric self-renewal divisions would be the most likely explanation of the faster rate of expansion in vivo of fetal HSCs.
Fig. 2.
Kinetics of HSC self-renewal divisions in vitro. (A) Schema showing the general experimental design used to determine the in vitro division kinetics of purified HSCs from E14.5 fetal liver. (B) Cumulative first (2 cells, pink symbols), second (≥3 cells, red symbols), and third divisions (≥5 cells, black symbols) in 80 cultures, each initiated with single purified E14.5 fetal liver HSC and maintained in 50 ng/ml SF only (data pooled from two experiments). Shown for reference in the gray dotted lines are corresponding curves on the same scale for adult bone marrow HSCs stimulated with 300 ng/ml SF plus 20 ng/ml interleukin-11 plus 1 ng/ml Flt3-ligand to achieve similar maintenance of functional HSC activity (redrawn from ref. 5).
Quantitative Differences in the Lineage Outputs of Transplanted HSCs from Different Stages of Development.
We also analyzed the various types of mature leukocytes present after 16 weeks in the blood of mice transplanted with the HSCs from different sources. From these measurements, we found that the percentage of donor-derived WBCs that were granulocytes and monocytes (Ly6g+ and/or Mac1+ cells) was >2× as high in recipients of E14.5 fetal liver HSCs than in recipients of adult bone marrow HSCs (Fig. 1 E and F). A similarly increased proportion of donor-derived cells that were myeloid was seen in recipients of HSCs from E18.5 fetal bone marrow or 3-week postnatal bone marrow, and from secondary recipients of HSCs harvested from primary mice 1–2 weeks after the primary mice had been transplanted with E14.5 fetal liver HSCs. In contrast, a lower proportion of myeloid cells was seen in the leukocytes produced by HSCs obtained from 4-week-old donors (or their regenerated HSC progeny), as well as from HSCs that were >4 weeks older than those from E14.5 fetal livers. Interestingly, an output of myeloid cells intermediate between that exhibited by fetal and adult HSCs was seen in secondary recipients of HSCs from primary mice that had been engrafted with 3-week postnatal bone marrow cells for just 1 week, suggesting that the HSCs present at that time comprised a mixture of HSCs with fetal and adult properties.
Changes in Gene Expression Parallel the Switch in Biological Properties of Juvenile HSCs.
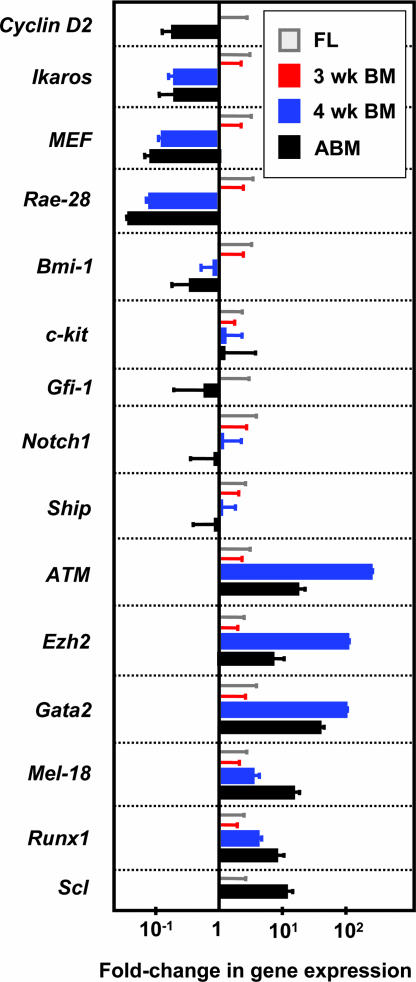
Collectively, these findings point to the operation of an intrinsically timed switch that simultaneously and abruptly alters the self-renewal and lineage restriction properties of HSCs (or the postrestriction amplification properties of their differentiating progeny) 5–6 weeks after the first HSCs appear in the embryo on E9 (9). It was, therefore, of interest to determine whether this switch in the biological properties of HSCs is accompanied by a change in their gene expression profile. To test this possibility, we isolated phenotypically defined populations of E14.5 fetal liver and adult bone marrow that are highly enriched in HSCs [20% pure (SI Fig. 5 A and D) and 40% pure (5), respectively]. Quantitative real-time PCR (Q-RT-PCR) measurements of the relative levels of transcripts for several genes revealed that some (Bmi-1, c-Kit, Gfi-1, Notch1, and Ship) were expressed at the same level in both of these HSC-enriched, albeit phenotypically distinct populations, with other genes showing a differential pattern of expression. The latter included genes reported to promote HSC cycling and/or self-renewal [CyclinD2, Ikaros, MEF, and Rae-28 (19–22)] that were more highly expressed in the fetal HSCs and others likely to be involved in HSC maintenance [ATM, Ezh2, Gata-2, and Mel-18 (20, 22, 23)] that were more highly expressed in adult HSCs (Fig. 3). The expression of two genes thought to be critical to the generation of HSCs in the fetus but not in the adult [Runx (24) and Scl (25, 26)] were up-regulated in the populations enriched in adult as compared with fetal HSCs. However, the expression of Notch1, which has also been ascribed a similar differential activity (27), was not different between the two populations.
Fig. 3.
Evidence of a switch during ontogeny in the gene expression profile of HSCs. Relative transcript levels were determined by comparing expression values obtained for lin− Rho− SP cells isolated from adult bone marrow (ABM, black bars) to those obtained for lin− Sca-1+ CD43+ Mac1+ cells isolated from E14.5 fetal liver (FL, values set = 1, gray bars) after internal normalization to Gapdh transcript levels. A similar comparison was made for extracts of the same populations isolated from 3- and 4-week postnatal bone marrow cells [3-wk BM (red bars) values set = 1, and 4-wk BM (blue bars), respectively]. Results are the mean ± SEM of data from two to three biological replicates, each measured in triplicate.
Preliminary experiments established that the same two strategies for obtaining highly enriched populations of fetal and adult HSCs allowed similarly enriched populations of transplantable HSCs to be obtained from the bone marrow of 3-week-old mice (SI Fig. 5B) and 4-week-old mice (SI Fig. 5C), respectively; i.e., 15% of the lin− Sca-1+ CD43+ Mac1+ bone marrow cells from 3-week-old mice and ≈25% of the lin− Rho− side population (SP) bone marrow cells from 4-week-old mice were found to be HSCs by limiting dilution transplantation assays (data not shown). Assessment of the transcripts present in these purified neonatal bone marrow cells showed a pattern of preserved and differential gene expression remarkably similar to that seen when fetal and adult HSC-enriched populations were compared (Fig. 3). However, these findings must be interpreted with caution given that different methods were used to enable similar purities of cycling and quiescent HSCs to be achieved. Nevertheless, it is interesting to note that the difference in expression of ATM, Ezh2, and Gata-2 in the HSCs-enriched cells between 3 and 4 weeks after birth was much more marked than the difference seen when analogous HSC-enriched populations of fetal liver and adult bone marrow cells were compared. It is thus conceivable that the products of these three genes, or regulators of their expression, may play an important role in the abrupt reprogramming of HSCs that occurs in the fourth week after birth (Fig. 4). Gata-2 may be of particular interest in this regard because its expression in fetal and adult HSCs is driven by different promoters (20, 23).
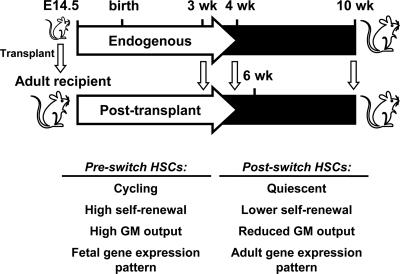
Fig. 4.
HSCs show a coordinated reprogramming of multiple key properties. Preswitch HSCs are actively cycling and demonstrate a high rate of self-renewal after transplantation into irradiated hosts, a higher output of myeloid (GM) cells, and a distinct gene expression profile. Postswitch HSCs are predominantly quiescent and display a lower rate of self-renewal after transplantation into irradiated hosts, a lower proportional output of myeloid cells, and a different gene expression profile from preswitch HSCs.
In summary, we have identified evidence of a previously unrecognized biologic switch that abruptly and rapidly alters several important properties of normal HSCs during ontogeny and appears intrinsically determined. Preliminary analysis of the molecular mechanism(s) involved (18) indicates that it affects a pathway affected by c-kit, a key receptor in mouse HSC self-renewal control and one whose activation differentially regulates fetal and adult HSC self-renewal both in vivo and in vitro (12, 28). The tight coordination seen in the alteration of multiple key HSC properties suggests the operation of a mechanism that may be relevant to understanding the regulation of other normal or perturbed stem cell types (29).
Methods
Cells.
Single-cell suspensions were prepared from the livers and bone marrow of embryos and 3- to 12-week-old C57BL/6J-Ptprca Pep3b/BoyJ (Peb3b-CD45/Ly5.1) mice as described (15). In some cases, Ter119+ cells were removed immunomagnetically from E14.5 fetal liver cells (using EasySep, StemCell Technologies, Vancouver, BC, Canada). Highly enriched HSC suspensions were obtained from E14.5 fetal liver and 3-week postnatal bone marrow cells by FACS selection of propidium iodide-negative (PI−) cells that stained positively after labeling with phycoerythrin (PE)-conjugated anti-ScaI, fluorescein isothiocyanate-conjugated anti-CD43, and allophycocyanin-conjugated anti-Mac1 (all from Becton Dickinson, San Jose, CA) and stained negatively after labeling with lineage-specific (lin) antibodies (using streptavidin-conjugated PE-Texas Red to detect biotinylated anti-Gr-1, anti-B220, and anti-Ly1, all prepared in the Terry Fox Laboratory; and Ter-119, anti-CD4, and anti-NK1.1, all from Becton Dickinson). Highly enriched HSC suspensions were obtained from 4- and 10-week postnatal bone marrow cells by FACS selection of lin− Rho− SP cells as described (5). Cells were sorted on a FACSVantage (Becton Dickinson).
HSC Transplantation and Quantification.
C57BL/6J-W41/W41 (CD45-Ly5.2) mice were irradiated with 360 cGy of 250-kVp x-rays, and then varying numbers of Peb3b-CD45/Ly5.1 cells were injected as indicated. Peripheral blood samples were obtained 16 weeks later, and the leukocytes were stained with anti-Ly5.1 and anti-Ly5.2 antibodies, as well as anti-B220, anti-Ly1, anti-Ly6g, and anti-Mac1 antibodies. Mice were considered to have been repopulated by at least one transplanted HSC if the WBCs contained ≥1% Ly5.1+ cells, including a ≥1% contribution of Ly5.1+ cells to each of the B220+ (B cell), Ly1+ (T cell), and Ly6g+/Mac1+ (myeloid) lineages. In most cases, HSC frequencies were calculated by using Poisson statistics and the method of maximum likelihood (L-Calc software; StemCell Technologies) from the proportions of mice injected with varying doses of cells that were considered negative by these criteria (30). When single-cell transplants of highly enriched HSC subsets were performed (for details of this protocol, see ref. 5), the HSC frequency was calculated directly from the number of positive recipients obtained. In the case of cycling HSCs from E14.5 fetal liver and 3-week postnatal bone marrow, the measured HSC purities can be assumed to represent underestimates by a factor of ≈2 because HSCs in S/G2/M cannot be detected (15). The total number of donor-derived HSCs regenerated per recipient was calculated assuming the bone marrow HSC content of two femurs and two tibias represents ≈25% (31).
Short-Term Cultures.
The single-cell deposition unit of the FACSVantage was used to deliver single lin− Sca-1+ CD43+ Mac1+ E14.5 fetal liver cells directly into the individual round bottom wells of a 96-well plate preloaded with Iscove's medium, a serum substitute (BIT; StemCell Technologies), 10−4 mM 2-mercaptoethanol, and 50 ng/ml Steel factor (SF) (StemCell Technologies). The presence of a single cell in each well was visually confirmed, the plate was then incubated at 37°C for up to 50 h, and each well was repeatedly examined by using an inverted microscope to determine the timing of the first, second, and third cell divisions (the interval until two, then at least three, and, finally, at least five cells were first seen in each well).
Gene Expression Analyses.
RNA was extracted by using the PicoPure RNA Isolation Kit (Arcturus Biosciences) by using a 15-min column DNase1 treatment (Qiagen) and was then eluted into an 11-μl volume and stored at −80°C. cDNA preparations were generated by using the SuperScript III First-Strand Synthesis System for RT-PCR (18080093; Invitrogen, Carlsbad, CA), with the reaction scaled to 25 μl. Q-RT-PCR was performed by using the following primer pairs (5′ to 3′): ATM (NM_007499.1) forward GCAGAGTGTCTGAGGGTTTGT and reverse AACTTCCAGCAACCTTCACC; Bmi-1 (NM_007552.3) forward AAACCAGACCACTCCTGAACA and reverse TCTTCTTCTCTTCAT-CTCATTTTTGA; c-kit (NM_021099.2) forward GATCTGCTCTGCGTCCTGTT and reverse CTGATTGTGCTGGATGGATG; CyclinD2 (NM_009829.2) forward GGCCAAGATCA-CCCACACT and reverse ATGCTGCTCTTGACGGAACT; Ezh2 (NM_007971.1) forward CATCGAAGGCAGTGGAGTC and reverse GTCTGGCCCATGATTATTCTTC; Gata-2 (NM_008090.3) forward TGACTATGGCAGCAGTCTCTTC and reverse ACACACTCCCGGCCTTCT; Gfi-1 (NM_010278.1) forward CTGCTCATTCACTCGGACAC and reverse ATTTGTGGGGCTTCTCACCT; Ikaros (NM_001025597) forward CCTGA-GGACCTGTCCACTACC and reverse ACGCCCATT-CTCTTCATCAC; MEF (NM_019680) forward TCTGTGGATGAGGAGGTTCC and reverse GGGTGCTGGAGAAGAA-CTCA; Mel-18 (NM_009545.1) forward TTCCCCCTCTTAACGATTTG and reverse GATCCTGGAGGCTGTTTCCT; Notch-1 (NM_008714.2) forward GCACAACTCCACTGATCCTG and reverse GCAAAGCCGACTTGCCTA; Rae-28 (NM_007905.1) forward GTCCCAGGCCCAGATGTAT and reverse CCCCATTAGGCATCAGGA; and Scl (NM_011527.1) forward TGAGATGGAGATTTCTGATGGTC and reverse CAAATGCCCCATTCACATT. Gapdh (NM_008084) was used as an endogenous control: forward AACTTTGGCATTGTGGAAGG and reverse ATGCAGGGATGATGTTCTGG.
Statistical Analyses.
Statistical comparisons were performed by using the Wald test except those in Fig. 1E, where Student's t test was used.
Supplementary Material
Acknowledgments
We thank the Flow Cytometry Facility of the Terry Fox Laboratory and the Animal Resource Centre of the British Columbia Cancer Agency, D. Wytrykush for secretarial assistance, P. Eirew for statistical advice, and C. Smith and A. Eaves for comments on the manuscript. This work was supported by the National Cancer Institute of Canada (with funds from the Terry Fox Foundation) (P.A.H., C.J.E., B.D.), the Stem Cell Network (M.B.B., D.G.K., B.D., C.J.E.), the National Heart, Lung, and Blood Institute/National Institutes of Health Grant P01 HL-55435 (to C.J.E.), the Canadian Institutes of Health Research and the Michael Smith Foundation for Health Research (M.B.B., D.G.K., P.A.H.), and the National Sciences and Engineering Research Council of Canada (K.D.M.).
Abbreviations
- CRUs
competitive repopulating units
- E
embryonic day
- HSC
hematopoietic stem cell
- lin
lineage markers
- PE
phycoerythrin
- PI
propidium iodide
- Q-RT-PCR
quantitative real-time PCR
- Rho
Rhodamine-123
- SF
steel factor
- SP
side population.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700460104/DC1.
References
- 1.Barnes DWH, Ford CE, Gray SM, Loutit JF. In: Progress in Nuclear Energy (Series VI) Bugher JC, Coursaget J, Loutit JF, editors. Oxford: Pergamon; 1959. pp. 1–10. [PubMed] [Google Scholar]
- 2.Keller G, Paige C, Gilboa E, Wagner EF. Nature. 1985;318:149–154. doi: 10.1038/318149a0. [DOI] [PubMed] [Google Scholar]
- 3.Lemischka IR, Raulet DH, Mulligan RC. Cell. 1986;45:917–927. doi: 10.1016/0092-8674(86)90566-0. [DOI] [PubMed] [Google Scholar]
- 4.Osawa M, Hanada KI, Hamada H, Nakauchi H. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 5.Uchida N, Dykstra B, Lyons KJ, Leung FYK, Eaves CJ. Exp Hematol. 2003;31:1338–1347. doi: 10.1016/j.exphem.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Szilvassy SJ, Nicolini FE, Eaves CJ, Miller CL. In: Methods in Molecular Medicine: Hematopoietic Stem Cell Protocols. Jordon CT, Klug CA, editors. Clifton, NJ: Humana; 2002. pp. 167–187. [DOI] [PubMed] [Google Scholar]
- 7.Gothert JR, Gustin SE, Hall MA, Green AR, Gottgens B, Izon DJ, Begley CG. Blood. 2005;105:2724–2732. doi: 10.1182/blood-2004-08-3037. [DOI] [PubMed] [Google Scholar]
- 8.Cheshier SH, Morrison SJ, Liao X, Weissman IL. Proc Natl Acad Sci USA. 1999;96:3120–3125. doi: 10.1073/pnas.96.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dzierzak E. Immunol Rev. 2002;187:126–138. doi: 10.1034/j.1600-065x.2002.18711.x. [DOI] [PubMed] [Google Scholar]
- 10.Harrison DE, Astle CM. J Exp Med. 1982;156:1767–1779. doi: 10.1084/jem.156.6.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rebel VI, Miller CL, Eaves CJ, Lansdorp PM. Blood. 1996;87:3500–3507. [PubMed] [Google Scholar]
- 12.Miller CL, Rebel VI, Helgason CD, Lansdorp PM, Eaves CJ. Blood. 1997;89:1214–1223. [PubMed] [Google Scholar]
- 13.Szilvassy SJ, Ragland PL, Miller CL, Eaves CJ. Exp Hematol. 2003;31:331–338. doi: 10.1016/s0301-472x(03)00005-5. [DOI] [PubMed] [Google Scholar]
- 14.Sudo K, Ema H, Morita Y, Nakauchi H. J Exp Med. 2000;192:1273–1280. doi: 10.1084/jem.192.9.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowie MB, McKnight KD, Kent DG, McCaffrey L, Hoodless PA, Eaves CJ. J Clin Invest. 2006;116:2808–2816. doi: 10.1172/JCI28310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eaves CJ, Eaves AC. In: Childhood Leukemias. Pui C-H, editor. Cambridge, UK: Cambridge Univ Press; 2006. pp. 69–105. [Google Scholar]
- 17.Pawliuk R, Eaves C, Humphries RK. Blood. 1996;88:2852–2858. [PubMed] [Google Scholar]
- 18.Bowie MB, Kent DG, Copley MR, Eaves CJ. Blood. 2007 doi: 10.1182/blood-2006-08-037770. in press. [DOI] [PubMed] [Google Scholar]
- 19.Sasaki Y, Jensen CT, Karlsson S, Jacobsen SE. Blood. 2004;104:986–992. doi: 10.1182/blood-2003-07-2277. [DOI] [PubMed] [Google Scholar]
- 20.Orkin S. Curr Opin Cell Biol. 1995;7:870–877. doi: 10.1016/0955-0674(95)80072-7. [DOI] [PubMed] [Google Scholar]
- 21.Lacorazza HD, Yamada T, Liu Y, Miyata Y, Sivina M, Nunes J, Nimer SD. Cancer Cell. 2006;9:175–187. doi: 10.1016/j.ccr.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 22.Kamminga LM, de Haan G. Stem Cells. 2006;24:1143–1149. doi: 10.1634/stemcells.2005-0345. [DOI] [PubMed] [Google Scholar]
- 23.Ling KW, Ottersbach K, van Hamburg JP, Oziemlak A, Tsai FY, Orkin SH, Ploemacher R, Hendriks RW, Dzierzak E. J Exp Med. 2004;200:871–882. doi: 10.1084/jem.20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ichikawa M, Asai T, Saito T, Seo S, Yamazaki I, Yamagata T, Mitani K, Chiba S, Ogawa S, Kurokawa M, Hirai H. Nat Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 25.Robb L, Elwood NJ, Elefanty AG, Kontgen F, Li R, Barnett LD, Begley CG. EMBO J. 1996;15:4123–4129. [PMC free article] [PubMed] [Google Scholar]
- 26.Curtis DJ, Hall MA, Van Stekelenburg LJ, Robb L, Jane SM, Begley CG. Blood. 2004;103:3342–3348. doi: 10.1182/blood-2003-09-3202. [DOI] [PubMed] [Google Scholar]
- 27.Hadland BK, Huppert SS, Kanungo J, Xue Y, Jiang R, Gridley T, Conlon RA, Cheng AM, Kopan R, Longmore GD. Blood. 2004;104:3097–3105. doi: 10.1182/blood-2004-03-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikuta K, Weissman IL. Proc Natl Acad Sci USA. 1992;89:1502–1506. doi: 10.1073/pnas.89.4.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rando TA. Nature. 2006;441:1080–1086. doi: 10.1038/nature04958. [DOI] [PubMed] [Google Scholar]
- 30.Dykstra B, Ramunas J, Kent D, McCaffrey L, Szumsky E, Kelly L, Farn K, Blaylock A, Eaves C, Jervis E. Proc Natl Acad Sci USA. 2006;103:8185–8190. doi: 10.1073/pnas.0602548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boggs DR. Am J Hematol. 1984;16:277–286. doi: 10.1002/ajh.2830160309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.