Abstract
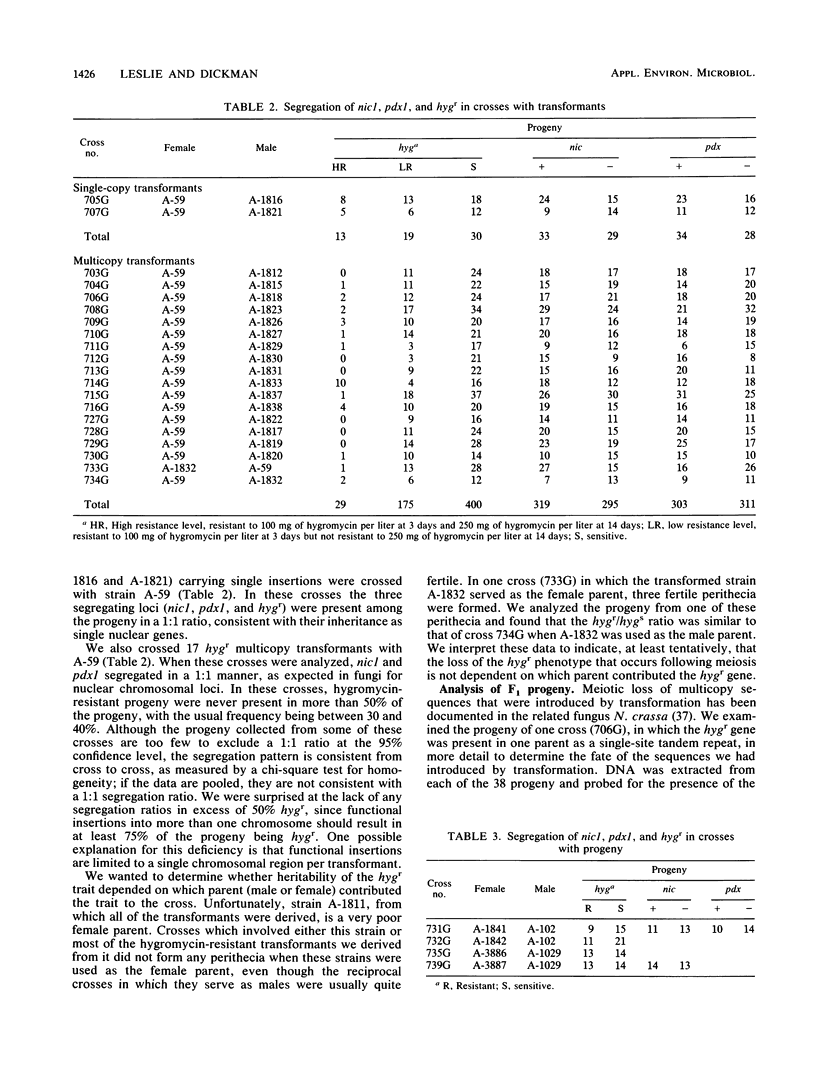
Stability of foreign DNA transformed into a novel host is an important parameter in decisions to permit the release of genetically engineered microorganisms into the environment. Meiotic instability of transformed DNA has been reported in fungi such as Ascobolus, Aspergillus, and Neurospora. We used strains of Gibberella fujikuroi (Fusarium moniliforme) transformed with the hygr gene from Escherichia coli to study meiotic stability of foreign DNA in this plant pathogenic fungus. Crosses with single-copy transformants segregated hygr:hygs in a 1:1 manner consistent with that expected for a Mendelian locus in a haploid organism. Multicopy transformants, however, segregated hygr:hygs in a 1:2 manner that was not consistent with Mendelian expectations for a chromosomal marker, even though two unrelated auxotrophic nuclear genes were segregating normally. Segregation ratios in crosses in which hygr was introduced via the male parent did not differ significantly from crosses in which the transformed strain served as the female parent. Some of the sensitive progeny from the crosses with the multicopy transformants carried hygr sequences. When these phenotypically sensitive progeny were crossed with a wild-type strain that carried no hygr sequences, some of the progeny were phenotypically hygr. Some progeny from some crosses were more resistant to hygromycin than were their sibs or the transformant strains that served as their parents. Transformants passaged through a maize plant only rarely segregated progeny with the high levels of resistance. The mechanism underlying these genetic instabilities is not clear but may involve unequal crossing over or methylation or both. Further work with cloned genes with homology to sequences already present in the Fusarium genome is warranted.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avalos J., Casadesús J., Cerdá-Olmedo E. Gibberella fujikuroi mutants obtained with UV radiation and N-methyl-N'-nitro-N-nitrosoguanidine. Appl Environ Microbiol. 1985 Jan;49(1):187–191. doi: 10.1128/aem.49.1.187-191.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case M. E. Genetical and molecular analyses of qa-2 transformants in Neurospora crassa. Genetics. 1986 Jul;113(3):569–587. doi: 10.1093/genetics/113.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincham J. R. Transformation in fungi. Microbiol Rev. 1989 Mar;53(1):148–170. doi: 10.1128/mr.53.1.148-170.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel S., Welch J. W., Maloney D. H. The molecular genetics of copper resistance in Saccharomyces cerevisiae--a paradigm for non-conventional yeasts. J Basic Microbiol. 1988;28(3):147–160. doi: 10.1002/jobm.3620280302. [DOI] [PubMed] [Google Scholar]
- Fogel S., Welch J. W. Tandem gene amplification mediates copper resistance in yeast. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5342–5346. doi: 10.1073/pnas.79.17.5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyon C., Faugeron G. Targeted transformation of Ascobolus immersus and de novo methylation of the resulting duplicated DNA sequences. Mol Cell Biol. 1989 Jul;9(7):2818–2827. doi: 10.1128/mcb.9.7.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R. K., Burlage R. S., Sayler G. S. Methods for detecting recombinant DNA in the environment. Crit Rev Biotechnol. 1988;8(1):33–84. doi: 10.3109/07388558809150537. [DOI] [PubMed] [Google Scholar]
- Kathariou S., Spieth P. T. Spore Killer Polymorphism in FUSARIUM MONILIFORME. Genetics. 1982 Sep;102(1):19–24. doi: 10.1093/genetics/102.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klittich C., Leslie J. F. Nitrate reduction mutants of fusarium moniliforme (gibberella fujikuroi). Genetics. 1988 Mar;118(3):417–423. doi: 10.1093/genetics/118.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malardier L., Daboussi M. J., Julien J., Roussel F., Scazzocchio C., Brygoo Y. Cloning of the nitrate reductase gene (niaD) of Aspergillus nidulans and its use for transformation of Fusarium oxysporum. Gene. 1989 May 15;78(1):147–156. doi: 10.1016/0378-1119(89)90322-3. [DOI] [PubMed] [Google Scholar]
- Perkins D. D., Radford A., Newmeyer D., Björkman M. Chromosomal loci of Neurospora crassa. Microbiol Rev. 1982 Dec;46(4):426–570. doi: 10.1128/mr.46.4.426-570.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rodriguez R. J., Yoder O. C. Selectable genes for transformation of the fungal plant pathogen Glomerella cingulata f. sp. phaseoli (Colletotrichum lindemuthianum). Gene. 1987;54(1):73–81. doi: 10.1016/0378-1119(87)90349-0. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Cambareri E. B., Jensen B. C., Haack K. R. Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell. 1987 Dec 4;51(5):741–752. doi: 10.1016/0092-8674(87)90097-3. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Garrett P. W. DNA sequence duplications trigger gene inactivation in Neurospora crassa. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6870–6874. doi: 10.1073/pnas.85.18.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan R. J., Atlas R. M. DNA amplification to enhance detection of genetically engineered bacteria in environmental samples. Appl Environ Microbiol. 1988 Sep;54(9):2185–2191. doi: 10.1128/aem.54.9.2185-2191.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilburn J., Scazzocchio C., Taylor G. G., Zabicky-Zissman J. H., Lockington R. A., Davies R. W. Transformation by integration in Aspergillus nidulans. Gene. 1983 Dec;26(2-3):205–221. doi: 10.1016/0378-1119(83)90191-9. [DOI] [PubMed] [Google Scholar]
- Turgeon B. G., Garber R. C., Yoder O. C. Development of a fungal transformation system based on selection of sequences with promoter activity. Mol Cell Biol. 1987 Sep;7(9):3297–3305. doi: 10.1128/mcb.7.9.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelton M. M., Timberlake W. E., Hondel C. A. A cosmid for selecting genes by complementation in Aspergillus nidulans: Selection of the developmentally regulated yA locus. Proc Natl Acad Sci U S A. 1985 Feb;82(3):834–838. doi: 10.1073/pnas.82.3.834. [DOI] [PMC free article] [PubMed] [Google Scholar]




