Abstract
We have cloned a human cDNA that is related to the RNA polymerase I transcription factor Rrn3 of Saccharomyces cerevisiae. The recombinant human protein displays both sequence similarity and immunological crossreactivity to yeast Rrn3 and is capable of rescuing a yeast strain carrying a disruption of the RRN3 gene in vivo. Point mutation of an amino acid that is conserved between the yeast and human proteins compromises the function of each factor, confirming that the observed sequence similarity is functionally significant. Rrn3 is the first RNA polymerase I-specific transcription factor shown to be functionally conserved between yeast and mammals, suggesting that at least one mechanism that regulates ribosomal RNA synthesis is conserved among eukaryotes.
Keywords: RRN3, ribosomal RNA, growth control, Saccharomyces cerevisiae
Eukaryotic ribosomal RNA (rRNA) genes are transcribed by an enzyme solely dedicated to that purpose, RNA polymerase I (pol I), and their expression is coordinated with cellular growth rate. When cell growth is impaired by nutrient deprivation or depletion, transcription of rRNA genes declines, and this decline is reversed when growth-permissive conditions are restored. Because growth-rate dependence is a universal feature of rRNA gene regulation, determining the molecular mechanisms coupling pol I activity to cell growth is a central question in studies of both yeast and mammalian systems.
Transcription of rRNA genes of Saccharomyces cerevisiae requires the activity of at least three transcription factors that have been both genetically and biochemically defined. Two of these are multisubunit factors that interact directly with distinct elements of the rDNA promoter to assemble a preinitiation complex. Core factor is composed of three essential gene products, RRN6, RRN7, and RRN11, associates with TATA-binding protein, and is required to direct initiation from the core promoter in vitro and in vivo (1–5). Upstream activation factor (UAF) binds to the upstream element and stimulates transcription from the core promoter. When the genes encoding the UAF subunits RRN5, RRN9, or RRN10 are individually disrupted, cells remain viable but exhibit pronounced growth defects, indicating that UAF activity is necessary to support levels of rRNA synthesis required for normal cell growth (6). UAF subunits interact with core factor subunits in vitro, and direct interaction of UAF with TATA-binding protein has been shown to mediate transcriptional activation in vivo (3, 7). The third transcription factor, Rrn3, is unique in that it functions as a single subunit, shows no sequence-specific DNA-binding activity, and is not required for preinitiation complex assembly (8). Rrn3 appears instead to function by direct interaction with RNA polymerase because it is stably associated with pol I in transcriptionally active extracts (9), and its transcription activity is enhanced by preincubation with pol I in the absence of either DNA template or other pol I transcription factors (4, 8). Interestingly, the interaction of Rrn3 with RNA polymerase fluctuates with changes in cellular growth rate: Rrn3 is not associated with pol I in transcriptionally inactive extracts prepared from growth-arrested cells, and activity is restored by addition of Rrn3-associated pol I purified from growing cells (9). These observations suggest that Rrn3 may be regulated in a growth-dependent manner. Although the specific function of Rrn3 is as yet unknown, it is required for rRNA gene transcription in vivo and in vitro, and it may mediate productive interactions of pol I with the preinitiation complex.
Transcription of mammalian rRNA genes also requires two promoter-binding transcription factors that appear to perform functions similar to those of the yeast pol I factors but whose protein components are not conserved with those of yeast. The core promoter-binding factor TIF-IB/SL1, which is essential for transcription, is comprised of TATA-binding protein and three associated factors that are similar in size to the three yeast core factor subunits but that display no sequence similarity to their yeast counterparts (10–12). The upstream stimulatory activity of mammalian cells is mediated by a single protein, upstream binding factor (UBF), which bears no resemblance to the multisubunit yeast UAF complex (13, 14). DNA binding by UBF is mediated by high mobility group domains that are not present in any of the yeast pol I transcription factors, and no proteins related to the yeast UAF subunits have been identified in mammals to date. It therefore appears that the promoter-binding factors of yeast and mammals have diverged extensively. Two RNA polymerase-associated factors, TIF-IA and TIF-IC, have been identified in mammalian cells (15, 16). Like yeast Rrn3 (yRrn3), TIF-IA and TIF-IC are not required for preinitiation complex assembly but are essential for transcription initiation (14). The relationship of these factors to Rrn3 has not yet been determined because their genes have not yet been isolated; however, TIF-IA shares an important functional similarity with Rrn3 in that its activity is regulated by cellular growth rate (15, 17).
In the present work, we describe the isolation of a human cDNA encoding a homolog of yeast RRN3 that displays both sequence similarity and immunological crossreactivity with the yeast transcription factor. Most importantly, the human cDNA rescues a lethal deletion of RRN3 when expressed in yeast. Unlike other pol I transcription factors, Rrn3 is functionally conserved between yeast and mammals.
Materials and Methods
Plasmid Constructs.
The yRRN3 ORF was PCR amplified from W3031a genomic DNA and cloned into pBluT (Novagen) to create RRN3-BluT. Four hundred seventy-six base pairs of 5′ sequence was amplified and inserted into RRN3-BluT to produce RRN3G. To express RRN3 from its own promoter, the RRN3G insert was subcloned into the PstI and BamHI sites of pRS425 (pRRN3G-425) or into the SpeI and BamHI sites of pRS316 (pRRN3G-316). Polyoma-tag expression constructs were generated by inserting a double-stranded oligonucleotide encoding two copies of the Py epitope and one copy of the FLAG tag into a pRS425 derivative containing the yeast PGK promoter (2). The ORFs of yRRN3, yeast mutant L143P, or hRRN3 were PCR amplified and inserted in-frame with the epitope cassette to create yPyWT, yPyL143P, and PyhRRN3. PyhL136P was generated by site-directed mutagenesis as described in ref. 18 by using oligonucleotide (5′-AACAGTCTGTGCTGATACAGGATTACCAAGAAAAGCCAA-3′) to introduce a leucine-to-proline substitution at position 136 of PyhRRN3. Full-length yeast or human RRN3 ORFs were subcloned into pRSET (Invitrogen) to generate 6His-tagged proteins for Escherichia coli expression.
Isolation of an rrn3− Strain.
RRN3-BluT was digested with EcoRV and PmeI, blunt ends created by using Klenow fragment, and a blunt-end XhoI/BamHI fragment of yeast His3 was inserted, replacing base pairs 492 to 1407 of the RRN3 ORF. An SpeI fragment of this construct was used to transform W1665a/α to His+, creating strain RLY300. Sporulation and dissection of this strain yielded two viable and two inviable spores, and viable colonies were His−. His3 insertion at the RRN3 locus was confirmed by both PCR and Southern blot analysis. RLY300 was transformed with pRRN3G-425 to Leu+. After sporulation and dissection, His+ Leu+ transformants were isolated and designated RLY301. To obtain an rrn3− strain containing RRN3 on a counterselectable Ura3 vector (RLY302), pRRN3G-425 was exchanged for pRRN3G-316 by using standard genetic techniques. To create a Nomura plasmid-dependent rrn3− strain, RLY302 was transformed with pNOY-W [a derivative of pNOY103 (19) with the Ura marker disrupted by Trp 1 insertion; a gift of P. Aprikian, Fred Hutchinson Cancer Research Center, Seattle] and plated on galactose medium. Trp+ colonies were cultured in galactose-Trp and spread on galactose plates containing 5-FOA. The resultant yeast (RLY303) were Trp+ and inviable on glucose.
Cloning of hRRN3.
A partial cDNA clone of human RRN3 was isolated from Jurkat cell RNA by reverse transcription–PCR (RT-PCR). Primer h5′.2: (5′-TGATTGCAGCAAAAAGTTAACCACTGA-3′ amino acids 508–517) was used to generate cDNA from 1 μg of total RNA (gift of A. M. Hajjar, University of Washington, Seattle) by using Superscript reverse transcriptase (GIBCO/BRL). Subsequent PCR with Pfu (Stratagene) by using gene-internal primers hRACE3′0.1: (5′-CTATATCGCGACCTGATAAACATCTTTC-3′ amino acids 343–354) and h5′.2 produced a 496-bp product that was inserted into pCR-Blunt (Invitrogen) to create clone h3–2. The 5′ end of the human mRNA was obtained by using a 5′RACE kit (GIBCO). An oligonucleotide complementary to the 5′ region of clone h3–2 (h5′.3: 5′-CAAAGATGTTTATCAGGTCGCGATATAG) was used to prime cDNA synthesis from Jurkat RNA, the cDNA product was purified, tailed, and PCR amplified according to the manufacturer's instructions, and the PCR product was cloned into pCR-Blunt to produce construct h5-L. The 3′ region of the human RRN3 ORF was isolated by using an anchor oligonucleotide complementary to the 5′ region of the polyA tail (CLONTECH) to prime a reverse transcription reaction as described above. The resultant cDNAs were amplified by PCR by using gene-specific primer h3′.1: (5′-CTATATCGCGACCTGATAAACATCTTTC-3′) and the polyA anchor primer. These PCR products were secondarily amplified by using primers h3′.2: (5′-GGAAGCTTTTTGGCAAGAGCTAAATTTATTCCTC-3′) and an expressed sequence tag (EST) AA31933 primer (5′ GCGGATCCTCATTCAGCACTCATGTCTTCCCATACCTGATA-3′: stop codon underlined) to produce a ≈500-bp PCR product. Three independent clones of all PCR products were sequenced. The hRRN3 ORF was constructed by joining the three partial cDNA clones. Construct h5-L was digested with SacI, filled in with Klenow, cleaved with NruI, and the fragment inserted at the EcoRV and NruI sites of clone h3–2. The cDNA encoding the C-terminal region was then ligated at an internal HindIII site to generate the hRRN3 construct used for further study. The sequence of the hRRN3 ORF was confirmed and has been deposited in the GenBank database (accession no. AF227156).
Antibody Generation and E. coli Expression.
The C-terminal region of yRRN3 (amino acids 352–627) was cloned into pRSETC (Invitrogen) to create a 6His-tagged fusion protein. The construct was transformed into E. coli strain UBS520, cultured in TBG/M9 to A600 = 0.8, and protein expression was induced by addition of isopropyl-d-thiogalactoside. The ≈30-kDa recombinant protein was isolated 3 h after induction by using Talon affinity resin (CLONTECH). The purified protein was resolved by SDS/PAGE, and the excised gel slices were used for immunization (R & R Rabbitry, Stanwood, WA). Antisera were fractionated by addition of (NH4)2SO4 to 50% saturation, the precipitate collected by centrifugation at 18,000 × g for 20 min, resupended in TBS, and dialyzed against TBS overnight at 4°C. Western blots were performed as described in ref. 2 with a 1:2,000 dilution of antiserum. For the Western blot shown in Fig. 2, cells were grown as described above except that 50 ml of medium was cultured and cells were lysed by boiling in SDS/PAGE buffer. Proteins were visualized by using anti-His monoclonal antibody (Qiagen).
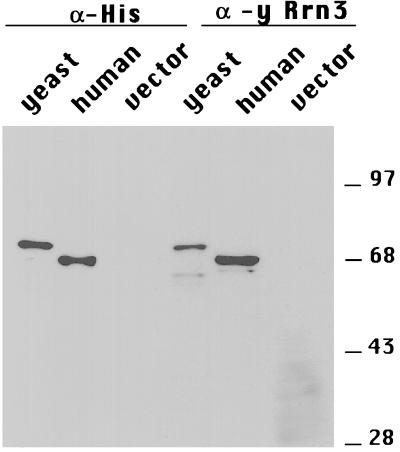
Figure 2.
Human Rrn3 crossreacts with anti-yRrn3 antibodies. E. coli-expressed 6His fusions of yeast and hRrn3 are shown. To compensate for differences in expression levels of the recombinant proteins, the volume of each lysate loaded was adjusted to permit equal amounts of yeast and hRrn3 to be compared. Nineteen microliters of yeast, 2 μl of human, or 19 μl of vector control lysate were resolved by SDS/PAGE and probed with monoclonal anti-6His antibodies (lanes 1–3) or with anti-yRrn3 antiserum (lanes 4–6). Positions of protein molecular weight markers are indicated.
Yeast Lysates.
Yeast transformants of Fig. 4 were grown to OD 600 = 2 in 50 ml of galactose medium. Cells were harvested by centrifugation, washed once with water, once with YEB (100 mM Hepes pH 7.9/20% glycerol/0.5 M KCl/5 mM EGTA/1 mM EDTA), and resuspended in 1.5 pellet volume of YEB containing 2.5 mM DTT and protease inhibitors (2). An equal volume of glass beads was added, and cells were lysed by shaking 4 × 40 sec in a Fastprep apparatus (Savant). The lysate was transferred and microfuged 20 min at 4°C. Fusion proteins were visualized by Western blot analysis by using anti-Py monoclonal antibody (20) at a 1/5,000 dilution.
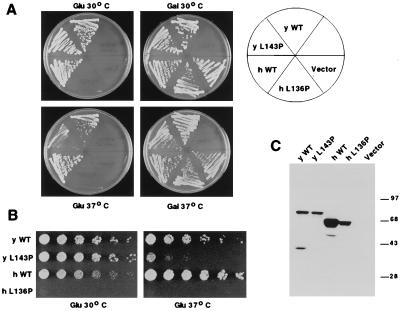
Figure 4.
Functional comparison of yeast L143P and human L136P mutants. (A) Transformants of rrn3− strain RLY303 expressing polyoma-tagged wild-type yRrn3 (yWT), yeast mutant L143P (yL143P), wild-type hRrn3 (hWT), human mutant L136P (hL136P), or empty expression vector are compared. Colonies were streaked on glucose to assay rrn3 complementation or onto galactose as a positive control and were incubated at 30°C or 37°C for 3 days. (B) Five-fold serial dilutions of the yeast and hRrn3 transformants compared in A were spotted onto glucose and incubated for 3 days at 30°C or 37°C. (C) Western blot monitoring expression of Rrn3 proteins in RLY303 transformants. Crude lysates were prepared from cells grown to OD 600 = 2.0 at 30°C in galactose media lacking leucine to select for the expression construct. Equal amounts of protein were loaded in each lane, and the blot was probed with a monoclonal antibody against the polyoma epitope. The identity of each expression construct is indicated above the lanes, and the positions of molecular weight standards are indicated to the right of the figure.
Database Searches.
Querying the available databases with the yRrn3 protein sequence in a blast search (21) uncovers full-length homologs in Schizosaccharomyces pombe [sp Q10110], Caenorhabditis elegans [sp P48322], and three paralogs in Arabidopsis thaliana [gb AAD25746, gb AAC16259, gb AAC28984]. Searching against unfinished genomic sequences reveals a near full-length sequence from Candida albicans [gnl Stanford 5476] and a partial sequence from Drosophila melanogaster mapping to chromosome 2 region 40D [gb AC011757]. Rrn3 homologs in other eukaryotes are recognized by their similarity to these sequences. A partial list of Rrn3 ESTs from other organisms includes mouse [gb AA530643], zebra fish [gb AI816698], Botrytis cinerea [emb AL112233], rice [gb AA752612], aspen [gb AI164160], and poplar [gb AI166768].
Results
Isolation of a Human RRN3 cDNA.
A blast search of the human EST database by using the yRrn3 protein sequence identified six overlapping ESTs encoding a protein fragment with homology to amino acids 360 to 532 of yRrn3. Primers derived from the EST sequences were used to isolate a partial cDNA from Jurkat cell RNA by RT-PCR, and sequence analysis of the product confirmed that it was identical to the contiguous EST sequence. The nucleotide sequence of this ORF-internal fragment (encoding amino acids 343 to 519 of the human protein) was used to design primers to isolate further 5′ and 3′ sequence. 5′ RACE generated two cDNA products that apparently arise from alternatively spliced mRNAs. Both cDNAs share identical 5′ sequences including an initiator methionine preceded by an in-frame stop codon, indicating that they encode the N terminus of the protein and that they are transcribed from the same promoter. The longer product contains an additional 90 bp of sequence encoding amino acids 245 to 334; because this region shares homology with yRrn3, the longer cDNA was used for further analysis.
Because we were unable to clone the 3′ end of the human message by 3′ RACE, the partial human cDNA was used to search the human EST databank for further 3′ sequence. The longest EST identified [gb AA319333] encodes an additional 70 amino acids, which extends the sequence alignment of the human protein to within eight residues of the yeast C terminus. Because this EST does not contain a translational stop codon, we designed a primer that inserted a translational stop codon adjacent to the final conserved residue (glutamate 587). This primer was used in conjunction with an ORF-internal 5′ primer to amplify the sequence encoding the conserved C-terminal region of the human protein.
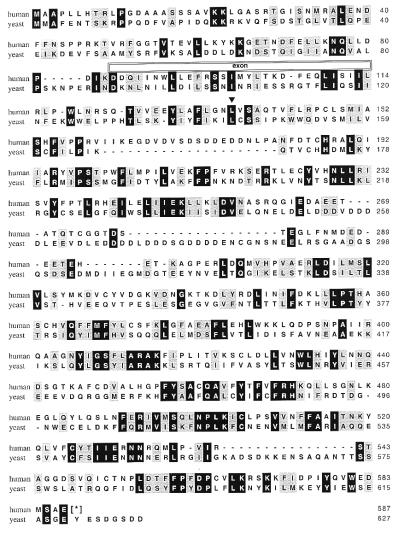
The resultant cDNA fragments were joined at overlapping restriction enzyme sites to produce an ORF encoding a 587-aa protein (Fig. 1). Although the low abundance of the human RRN3 message makes it difficult to detect by Northern blot analysis, we know that the cDNA represents a single gene product because, (i) RT-PCR by using the extreme 5′ and 3′ primers generates a product of the expected size and sequence (data not shown), and (ii) the cDNA sequence is contained within a genomic fragment of human chromosome region 16p12, which is found on two independent bacterial artificial chromosome clones (gb AF001549 and gb AC017077). Alignment of the cDNA and genomic sequences confirms that the 90-bp sequence present in the longer cDNA product represents an exon that is also present in chromosome 16. The predicted sequence of the human protein is 21% identical and 43% similar to yRrn3, and amino acid conservation is distributed throughout the length of the two proteins. Although no previously identified structural motifs are apparent in either protein, the human sequence contains a highly acidic region (amino acids 163–180), similar to that previously noted in yRrn3 (8), but which is located nearer the N terminus than is its yeast counterpart.
Figure 1.
Amino acid sequence alignment of human and yRrn3. Identical amino acids are highlighted on black, conservative substitutions on gray. The translational stop codon introduced by the 3′ primer used for RT-PCR is indicated by an asterisk. The position of an exon in the human cDNA is indicated. The arrowhead indicates the position of a leucine-to-proline substitution of a temperature-sensitive yeast RRN3 mutant.
Human and Yeast Rrn3 Are Antigenically Related.
To confirm that the human cDNA encodes a protein related to yRrn3, the human and yeast RRN3 cDNAs were expressed in E. coli as 6-His fusion proteins and subjected to Western blot analysis. Crude lysates prepared from bacteria expressing either the yeast or human cDNA or from cells transformed with the empty expression vector were resolved by SDS/PAGE and probed with an antibody against the His epitope or with an antiserum generated against the C-terminal half of recombinant yRrn3. The signal intensities obtained by using antibodies against the 6-His tag were used to normalize the amount of recombinant protein loaded. As shown in Fig. 2 (lanes 1–3), anti-His antibodies recognize proteins of 72 kDa and 68 kDa in lysates from cells expressing the yeast and human cDNAs, respectively, whereas no signal is observed in the lysate from the empty vector control. When these lysates were probed with an antiserum generated against yRrn3 (Fig. 2, lanes 4–6), we were surprised to observe that the human protein was more efficiently detected than the yeast protein. This may be due to limited proteolysis of yRrn3, because it is less stably expressed in bacteria than the human protein. However, no signal is observed in the vector control lysate, indicating that the human protein is specifically recognized by the anti-yRrn3 serum, and it is clear from the strength of the observed crossreaction signal that the yeast and human proteins are highly immunologically conserved.
Human RRN3 Complements an rrn3− Strain.
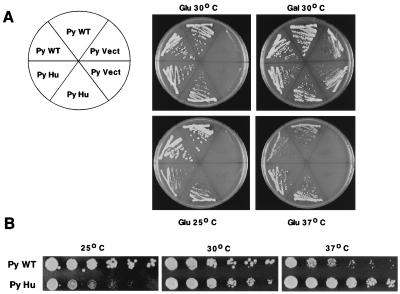
To determine whether human RRN3 was functionally related to the yeast transcription factor, we assayed its ability to rescue a yeast strain in which the genomic RRN3 allele was rendered nonfunctional by deletion and marker insertion. For this purpose, the human and yeast RRN3 ORFs were expressed as polyoma epitope-tagged fusion proteins from the yeast PGK promoter in rrn3− strain RLY303, which contains a plasmid expressing the 35S rRNA precursor under control of the GAL7 promoter. Because RRN3 is essential for rRNA gene transcription, this strain is viable only on galactose-containing media. RLY303 was transformed with either yeast wild-type RRN3, the human cDNA, or the empty expression vector and grown under appropriate selection on galactose plates. Transformants were then streaked on complete medium (CM)-glucose plates to monitor RRN3 function or on CM-galactose plates as a positive control and were incubated at 25°C, 30°C, or 37°C (Fig. 3A). When colonies are streaked onto glucose medium to repress transcription from the GAL7 promoter, cells expressing either yeast or human Rrn3 (hRrn3) grow at all temperatures tested, whereas those transformed with the empty expression construct are inviable. The ability of the human gene to complement the rrn3− mutation demonstrates that its function in pol I transcription is conserved between yeast and humans and that the region of the human protein encoded by the partial cDNA is sufficient for in vivo activity in yeast. Interestingly, the yeast and human RRN3 genes display distinct conditional phenotypes in the rrn3− background. This is most readily observed in Fig. 3B, in which serial 5-fold dilutions of cells transformed with either yeast or human RRN3 are compared. Cells carrying the yeast gene grow well at 25°C and 30°C but show slightly reduced growth at 37°C. In contrast, cells transformed with human RRN3 grow better than yeast RRN3 transformants at 37°C and show a pronounced reduction of growth at 25°C. These temperature-dependent growth differences are not observed when the transformed cells are maintained on galactose (data not shown).
Figure 3.
Human RRN3 complements the lethal phenotype of an rrn3− mutant. (A) rrn3− strain RLY303 was transformed with a 2-μm plasmid construct expressing polyoma-tagged yeast RRN3 (PyWT), human RRN3 (PyHu), or empty expression vector (PyVect). Two single colonies from each transformation were streaked on glucose (Glu) to assay rrn3 complementation or on galactose (Gal) as a positive control. Plates were incubated at the temperatures indicated for 3 days (30°C, 37°C) or 5 days (25°C) and photographed. (B) Effect of yeast or human RRN3 expression on cell growth. Five-fold serial dilutions of yeast (PyWT) or human (PyHu) RRN3 transformants were spotted onto glucose. Plates were incubated at 25°C, 30°C, or 37°C as described for A.
To determine whether the observed sequence conservation between yeast and hRrn3 contributes to its function, we compared the effect of a point mutation on the in vivo activity of the two proteins. We had previously isolated a temperature-sensitive mutant of yeast RRN3 that contained a leucine-to-proline substitution at position 143 and that showed reduced growth at 37°C (B.M. and R.H.R., unpublished work). Because protein sequence alignment indicated that this residue was conserved in hRrn3 (see Fig. 1), we introduced a proline at this position (L136P) by site-directed mutagenesis and compared the ability of the wild-type and mutant proteins to complement the rrn3− knockout strain (Fig. 4). The temperature-sensitive phenotype of the yeast L143P mutant is clearly observed when cells are streaked to glucose plates (Fig. 4A) or when serial dilutions of wild-type and mutant transformants are compared (Fig. 4B) at 30°C and 37°C. In contrast, the human RRN3 mutant L136P was unable to support growth on glucose at either temperature. To confirm that the mutant was stably expressed, lysates prepared from the transformants were subjected to Western blot analysis with an antipolyoma epitope antibody (Fig. 4C). Although full-length protein is readily detected in all transformants, the levels of the human L136P mutant are reduced with respect to the human wild-type protein. We therefore do not know whether increased expression of the hL136P mutant would result in a conditional phenotype similar to that observed with the yeast L143P mutant. However, we know from independent experiments that the loss of in vivo function of hL136P is not solely the result of reduced expression because the human wild-type protein is still capable of rescuing the rrn3− strain when it is expressed at levels comparable to those of hL136P (B.M. and R.H.R., unpublished data). Therefore, the conserved leucine residue contributes to the in vivo function of both yeast and hRrn3.
Discussion
We have identified a functional human homolog of the yeast pol I transcription factor Rrn3. The human cDNA, hRRN3, encodes a 587-aa protein that is 21% identical to yRrn3 (Fig. 1). Consistent with this amino acid conservation, recombinant hRrn3 is specifically recognized by an antiserum generated against yRrn3, indicating that the yeast and human proteins are immunologically related (Fig. 2). hRRN3 complements a lethal disruption of rrn3 when expressed in yeast, demonstrating that its function in pol I transcription has been conserved between yeast and humans and that the protein encoded by the partial human cDNA is sufficient for in vivo function (Fig. 3). Moreover, hRRN3 complements the rrn3 null strain nearly as well as yRRN3 at moderate temperatures and more strongly than yRRN3 at elevated temperatures. This high complementation efficiency was unexpected in light of the low sequence identity of the yeast and human proteins and suggests that Rrn3 activity is mediated by a discrete number of amino acids that have been strongly evolutionarily conserved.
Database searches reveal that the human cDNA is derived from a gene located on chromosome 16 that is >26 kb in length and that contains at least 15 introns. We have not examined the tissue distribution of hRRN3 because of difficulty visualizing the low-abundance message by Northern blot analysis. However, ESTs arising from the RRN3 gene have been isolated from a variety of tissue including lung, retina, thymus, and prostate, and ubiquitous expression of hRRN3 is consistent with its predicted role as a pol I transcription factor. Although the demonstration that hRrn3 regulates human rRNA gene transcription requires that its function be assayed in mammalian cells, its conservation between organisms as extensively diverged as humans and yeast suggests that it performs an essential function in eukaryotes. Furthermore, mammalian transcription factor TIF-IA closely resembles yRrn3; both factors associate with pol I, and their activities are growth rate dependent (9, 15, 17). Given these functional similarities, it is likely that hRrn3 is identical to TIF-IA.
The isolation of a human homolog of RRN3 facilitates the identification of RRN3-related genes in other organisms (Fig. 5). The emerging picture from database searches is that RRN3 is a member of a conserved family of genes that are found in organisms spanning three biological kingdoms: fungi (S. cerevisiae, S. pombe), animals (Homo sapiens, C. elegans), and plants (A. thaliana). Interestingly, three RRN3-related genes can be identified in this last organism.
Figure 5.
Comparison of Rrn3-related proteins. Alignment of the predicted protein sequences of Rrn3 from S. cerevisiae, S. pombe, H. sapiens, C. elegans, and A. thaliana. Amino acids that are identical or similar are highlighted in black or gray, respectively. Boxes labeled A, B, and C indicate three conserved regions whose function is unknown. The predicted PEST region of hRrn3 is double underlined. The arrowhead indicates the position of a conserved leucine residue that was mutated in the experiment shown in Fig. 4. Dashes indicate gaps introduced to maximize alignment.
Visual inspection of the cross-species sequence alignment reveals that conservation is biased toward the C-terminal half of the protein. Although the N-terminal region shows considerable sequence variation, we note that the leucine that was mutated in Fig. 4 (yL143P, hL136P) is conserved in all five organisms compared, supporting the observation that this residue is important for in vivo function. C-terminal sequence conservation is most striking in three regions of unknown function that are unique to Rrn3 proteins but that are shared by all Rrn3 family members. These regions are conserved in both sequence and length and display the general consensus: Y(I/L)(A/G)(A/S)(F/Y)(I/L)(A/S)RAK; FY(A/S)xxQ(A/S)(I/L)xxx (F)xFR; and FP(F/Y)DxxxL(K). As these motifs are not found in other proteins, their evolutionary conservation may reflect the pol I-specific function of Rrn3. Because yRrn3 does not bind DNA, these regions are likely to mediate protein–protein interactions with other components of the pol I transcriptional machinery. yRrn3 has been shown to interact with pol I subunits A49 and A34.5 in vitro (unpublished results cited in ref. 8) and stably associates with pol I in yeast extracts (9). These conserved motifs are therefore potentially interesting targets for future studies to investigate their contribution to Rrn3–pol I interaction.
Although the Rrn3-related proteins of fungi are conserved in length, the genes of C. elegans and A. thaliana are predicted to encode an extended C-terminal region that is observed only in multicellular organisms and that therefore may also be present in the human protein. If this is the case, the human cDNA that we have isolated encodes a “core” region that is shared by all RRN3 family members and that is sufficient for function in S. cerevisiae. The possible function of an additional C-terminal region, if any, must be addressed by future experiments.
Interestingly, residues 163–186 of hRrn3 comprise a predicted PEST region that is not present in the other proteins compared in the alignment. PEST sequences have been shown to be signals for regulated proteolysis of a wide variety of proteins, including several transcription factors (reviewed in ref. 22). It will therefore be of interest to investigate its contribution to hRrn3 stability in human cells and to see whether it is conserved in other mammalian Rrn3-related proteins as their sequences become available.
Although the function of hRrn3 remains to be determined, its ability to rescue a yeast rrn3− strain greatly facilitates the study of its activity. The yeast complementation assay can be used as a preliminary screen for mutations whose effects can be confirmed by studies in human cells. Yeast genetic screens can also be used to identify Rrn3-interacting proteins, which may in turn have human homologs. The ability to study Rrn3 function in both yeast and human cells expands the experimental approaches that can be used to determine the role of Rrn3 in transcription initiation and the factors that regulate its activity.
Growth-rate regulation of rRNA gene expression has been observed in organisms as evolutionarily diverse as yeast (9, 23), Acanthamoeba (24), and mammals (15, 17, 25–27). The identification of a human homolog of a yeast transcription factor that is regulated by growth rate (9), together with the identification of RRN3-related genes in a wide variety of organisms, raises the intriguing possibility that Rrn3-related factors may play a role in the regulation of rRNA transcription in other eukaryotes as well. Comparative studies of Rrn3 function in different organisms may therefore reveal aspects of a common mechanism regulating eukaryotic rRNA gene expression.
Table 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| W1665a/α | MATa/α ade2-1 his3-11 leu2-3,112 trp1-1,15 |
| ura3-1 can1-100 RAD5 | |
| RLY300 | MATa/α ade2-1 his3-11 leu2-3,112 trp1-1,15 |
| ura3-1 can1-100 RAD5 rrn3Δ∷HIS3 | |
| RLY301 | MATa ade2-1 his3-11 leu2-3,112 trp1-1,15 |
| ura3-1 can1-100 RAD5 rrn3Δ∷HIS3 [pRRN3G-425] | |
| RLY302 | MATa ade2-1 his3-11 leu2-3,112 trp1-1,15 |
| ura3-1 can1-100 RAD5 rrn3Δ∷HIS3 [pRRN3G-316] | |
| RLY303 | MATa ade2-1 his3-11 leu2-3,112 trp1-1,15 |
| ura3-1 can1-100 RAD5 rrn3Δ∷HIS3 [pNOY-TRP1] |
Acknowledgments
We thank A. M. Hajjar for human RNA samples and for advice on RT-PCR, P. Aprikian for pNOY-Trp, and J. Roan and J. Sporleder for excellent technical assistance. We also thank members of the S. Hahn and T. Tsukiyama labs for both reagents and helpful discussions. This work was partially supported by National Institutes of Health Grant GM26624 (awarded to R.H.R.). B.M. was supported by Training Grant CA09657 from the National Cancer Institute.
Abbreviations
- rRNA
ribosomal RNA
- pol I
RNA polymerase I
- UAF
upstream activation factor
- hRrn3
human Rrn3
- yRrn3
yeast Rrn3
- EST
expressed sequence tag
- RACE
rapid amplification of cDNA ends
Note Added in Proof
A cDNA encoding the C terminus of hRrn3 was cloned by using a primer derived from the recently reported human EST sequence gb AW239267. The full-length hRRN3 cDNA, which also complements the rrn3− strain, encodes a 651-aa protein with a 74-kDa predicted molecular mass similar to that of TIF-IA. The GenBank report has been updated to include the full-length sequence.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF227156).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080063997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080063997
References
- 1.Keys D A, Vu L, Steffan J S, Dodd J A, Yamamoto R T, Nogi Y, Nomura M. Genes Dev. 1994;8:2349–2362. doi: 10.1101/gad.8.19.2349. [DOI] [PubMed] [Google Scholar]
- 2.Lin C W, Moorefield B, Payne J, Aprikian P, Mitomo K, Reeder R H. Mol Cell Biol. 1996;16:6436–6443. doi: 10.1128/mcb.16.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steffan J S, Keys D A, Dodd J A, Nomura M. Genes Dev. 1996;10:2551–2563. doi: 10.1101/gad.10.20.2551. [DOI] [PubMed] [Google Scholar]
- 4.Keener J, Josaitis C A, Dodd J A, Nomura M. J Biol Chem. 1998;273:33795–33802. doi: 10.1074/jbc.273.50.33795. [DOI] [PubMed] [Google Scholar]
- 5.Lalo D, Steffan J S, Dodd J A, Nomura M. J Biol Chem. 1996;271:21062–21067. doi: 10.1074/jbc.271.35.21062. [DOI] [PubMed] [Google Scholar]
- 6.Keys D A, Lee B S, Dodd J A, Nguyen T T, Vu L, Fantino E, Burson L M, Nogi Y, Nomura M. Genes Dev. 1996;10:887–903. doi: 10.1101/gad.10.7.887. [DOI] [PubMed] [Google Scholar]
- 7.Steffan J S, Keys D A, Vu L, Nomura M. Mol Cell Biol. 1998;18:3752–3761. doi: 10.1128/mcb.18.7.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto R T, Nogi Y, Dodd J A, Nomura M. EMBO J. 1996;15:3964–3973. [PMC free article] [PubMed] [Google Scholar]
- 9.Milkereit P, Tschochner H. EMBO J. 1998;17:3692–3703. doi: 10.1093/emboj/17.13.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comai L, Tanese N, Tjian R. Cell. 1992;68:965–976. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- 11.Eberhard D, Tora L, Egly J M, Grummt I. Nucleic Acids Res. 1993;21:4180–4186. doi: 10.1093/nar/21.18.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heix J, Zomerdijk J C M B, Ravanpay A, Tjian R, Grummt I. Proc Natl Acad Sci USA. 1997;94:1733–1738. doi: 10.1073/pnas.94.5.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell S P, Learned R M, Jantzen H-M, Tjian R. Science. 1988;241:1192–1197. doi: 10.1126/science.3413483. [DOI] [PubMed] [Google Scholar]
- 14.Schnapp A, Grummt I. J Biol Chem. 1991;266:24588–24595. [PubMed] [Google Scholar]
- 15.Schnapp A, Pfleiderer C, Rosenbauer H, Grummt I. EMBO J. 1990;9:2857–2863. doi: 10.1002/j.1460-2075.1990.tb07475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnapp G, Schnapp A, Rosenbauer H, Grummt I. EMBO J. 1994;13:4028–4035. doi: 10.1002/j.1460-2075.1994.tb06719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnapp A, Schnapp G, Erny G, Grummt I. Mol Cell Biol. 1993;13:6723–6732. doi: 10.1128/mcb.13.11.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nogi Y, Vu L, Nomura M. Proc Natl Acad Sci USA. 1991;88:7026–7030. doi: 10.1073/pnas.88.16.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grussenmeyer T, Scheidtmann K H, Hutchinson M A, Eckhart W, Walter G. Proc Natl Acad Sci USA. 1985;82:7952–7954. doi: 10.1073/pnas.82.23.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rechsteiner M, Rogers S W. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 23.Riggs D L, Peterson C L, Wickham J Q, Miller L M, Clarke E M, Crowell J A, Sergere J C. J Biol Chem. 1995;270:6205–6210. doi: 10.1074/jbc.270.11.6205. [DOI] [PubMed] [Google Scholar]
- 24.Bateman E, Paule M R. Cell. 1986;47:445–450. doi: 10.1016/0092-8674(86)90601-x. [DOI] [PubMed] [Google Scholar]
- 25.Tower J, Sollner Webb B. Cell. 1987;50:873–883. doi: 10.1016/0092-8674(87)90514-9. [DOI] [PubMed] [Google Scholar]
- 26.Mahajan P B, Gokal P K, Thompson E A. J Biol Chem. 1990;265:16244–16247. [PubMed] [Google Scholar]
- 27.Mahajan P B, Thompson E A. J Biol Chem. 1990;265:16225–16233. [PubMed] [Google Scholar]