Abstract
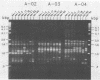
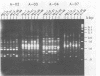
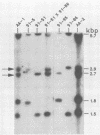
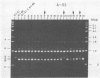
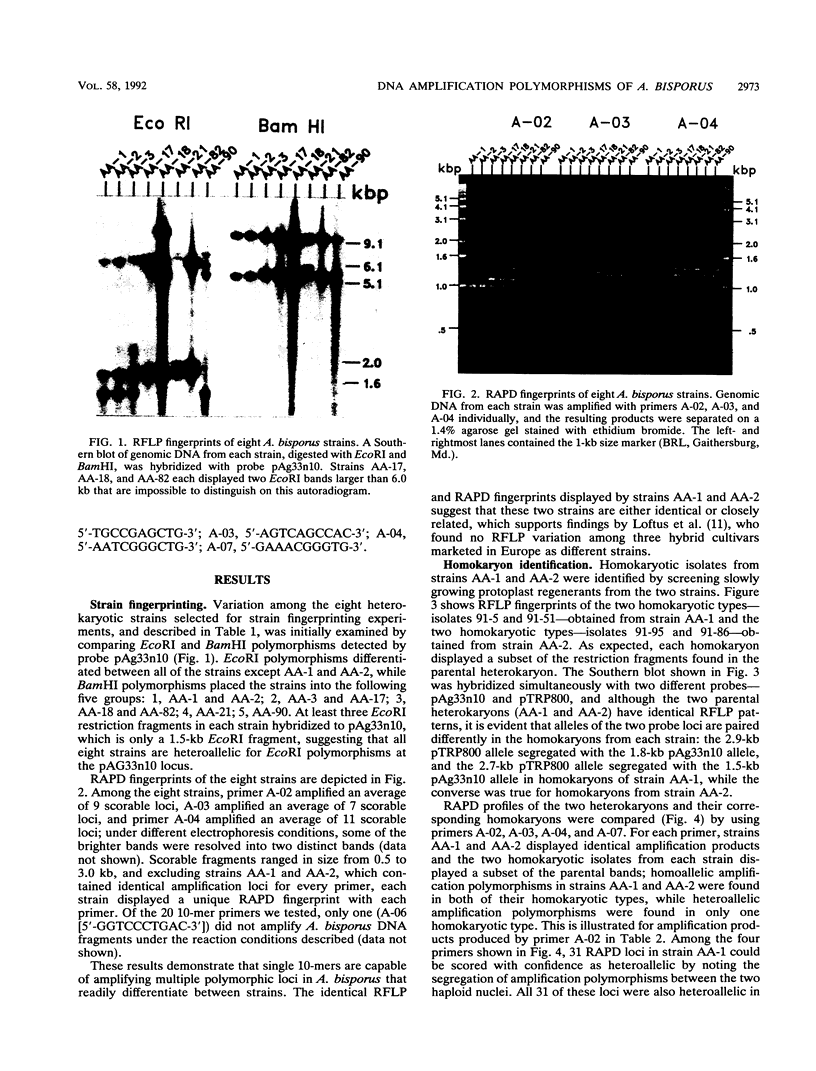
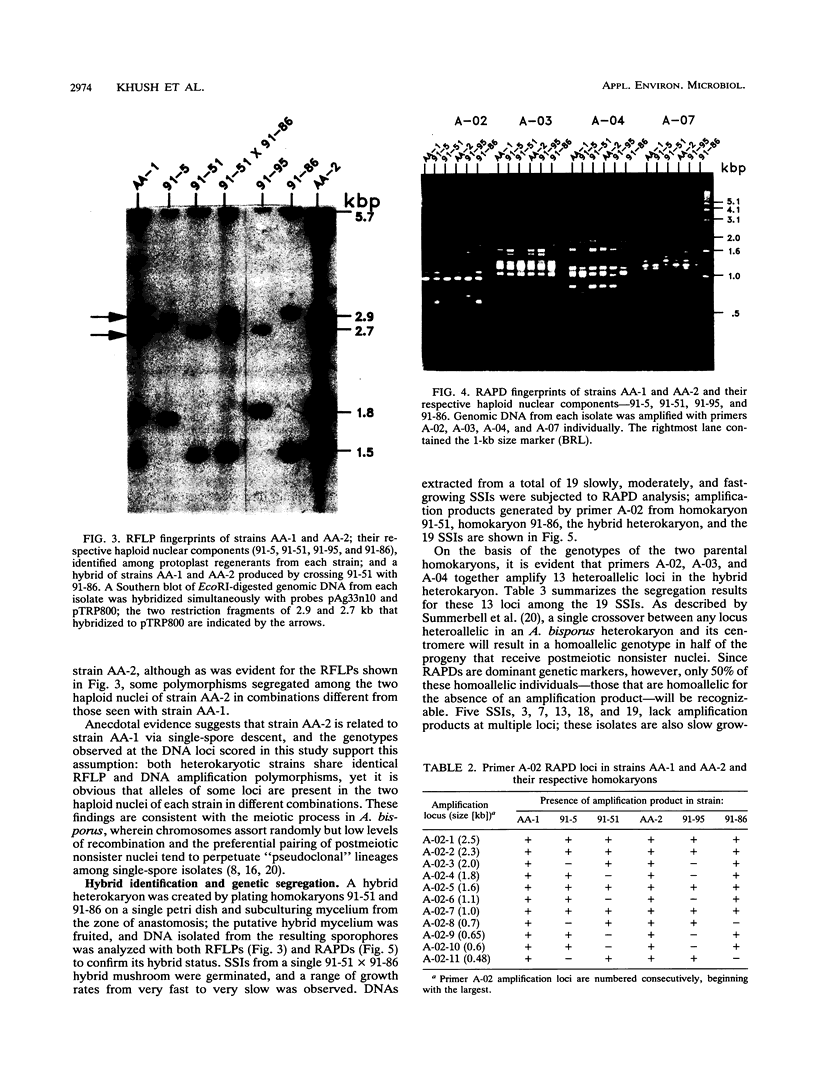
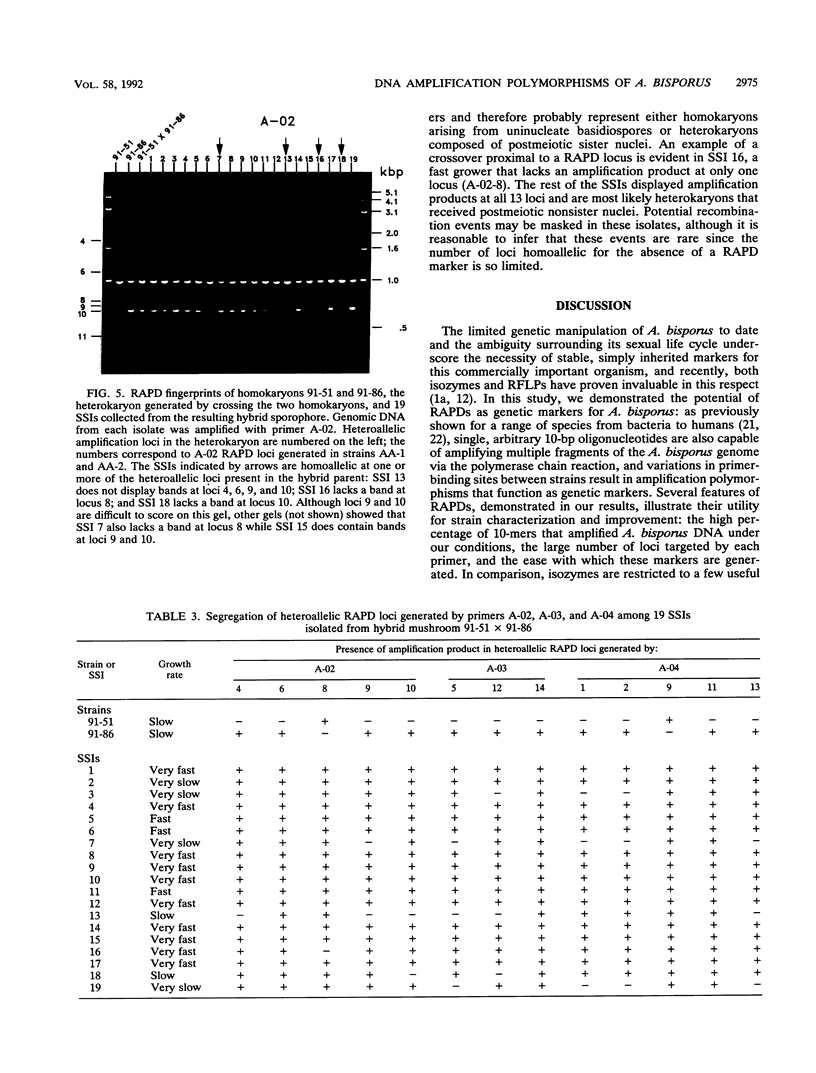
Single 10-bp primers were used to generate random amplified polymorphic DNA (RAPD) markers from commercial and wild strains of the cultivated mushroom Agaricus bisporus via the polymerase chain reaction. Of 20 primers tested, 19 amplified A. bisporus DNA, each producing 5 to 15 scorable markers ranging from 0.5 to 3.0 kbp. RAPD markers identified seven distinct genotypes among eight heterokaryotic strains; two of the commercial strains were shown to be related to each other through single-spore descent. Homokaryons recovered from protoplast regenerants of heterokaryotic strains carried a subset of the RAPD markers found in the heterokaryon, and both of the haploid nuclei from two heterokaryons were distinguishable. RAPD markers also served to verify the creation of a hybrid heterokaryon and to analyze meiotic progeny from this new strain: most of the basidiospores displayed RAPD fingerprints identical to that of the parental heterokaryon, although a few selected slow growers were homoallelic at a number of loci that were heteroallelic in the parent, suggesting that they represented rare homokaryotic basidiospores; crossover events between a RAPD marker locus and its respective centromere appeared to be infrequent. These results demonstrate that RAPD markers provide an efficient alternative for strain fingerprinting and a versatile tool for genetic studies and manipulations of A. bisporus.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Castle A. J., Horgen P. A., Anderson J. B. Crosses among Homokaryons from Commercial and Wild-Collected Strains of the Mushroom Agaricus brunnescens (= A. bisporus). Appl Environ Microbiol. 1988 Jul;54(7):1643–1648. doi: 10.1128/aem.54.7.1643-1648.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle A. J., Horgen P. A., Anderson J. B. Restriction fragment length polymorphisms in the mushrooms Agaricus brunnescens and Agaricus bitorquis. Appl Environ Microbiol. 1987 Apr;53(4):816–822. doi: 10.1128/aem.53.4.816-822.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EVANS H. J. Nuclear behaviour in the cultivated mushroom. Chromosoma. 1959;10(2):115–135. doi: 10.1007/BF00396566. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Summerbell R. C., Castle A. J., Horgen P. A., Anderson J. B. Inheritance of restriction fragment length polymorphisms in Agaricus brunnescens. Genetics. 1989 Oct;123(2):293–300. doi: 10.1093/genetics/123.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990 Dec 25;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]