Abstract
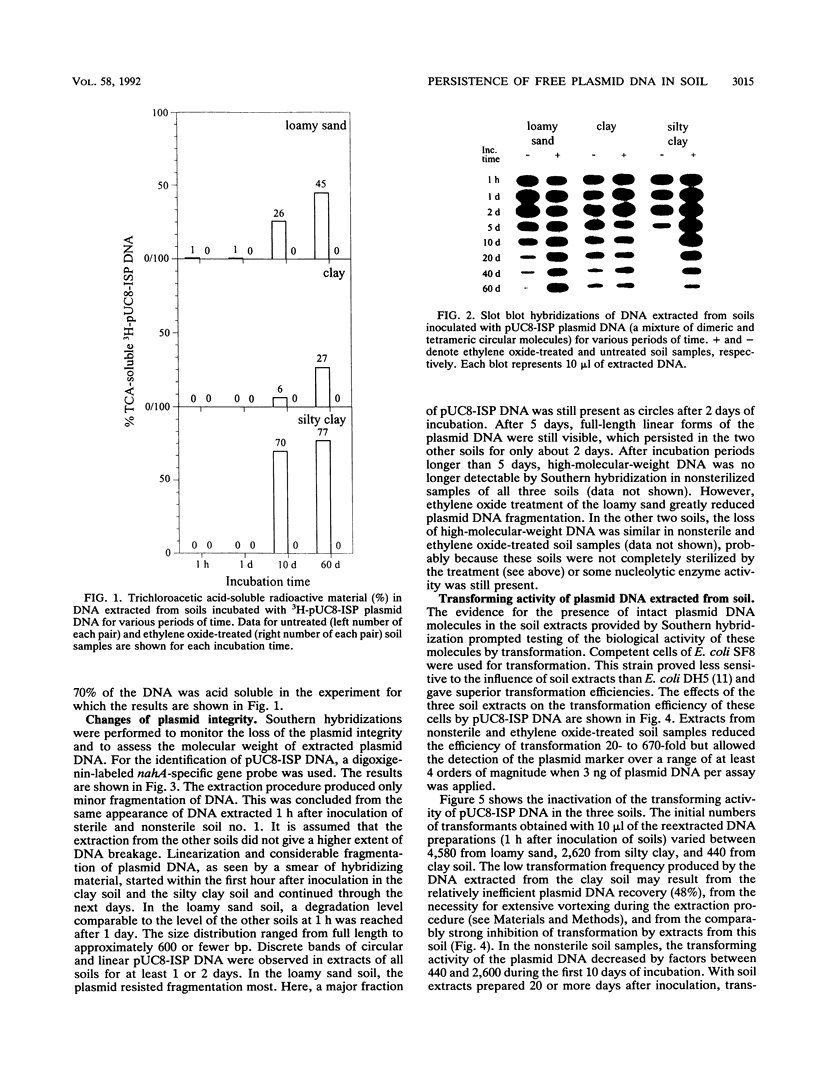
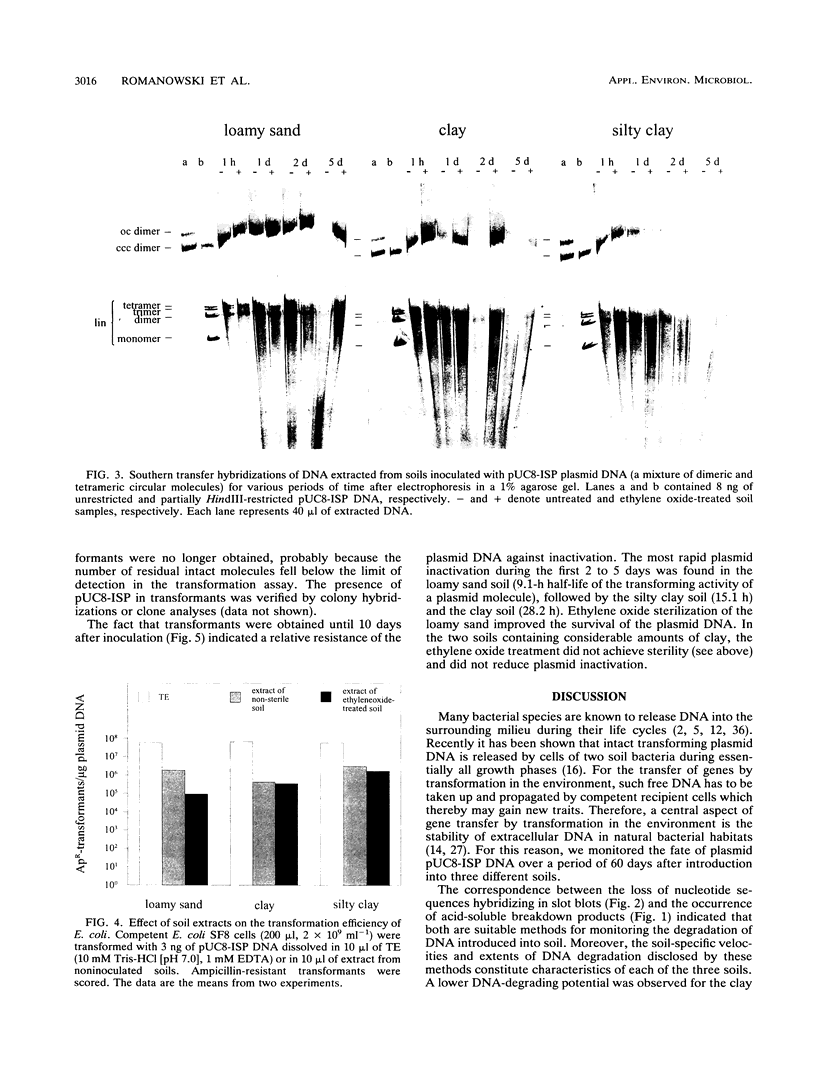
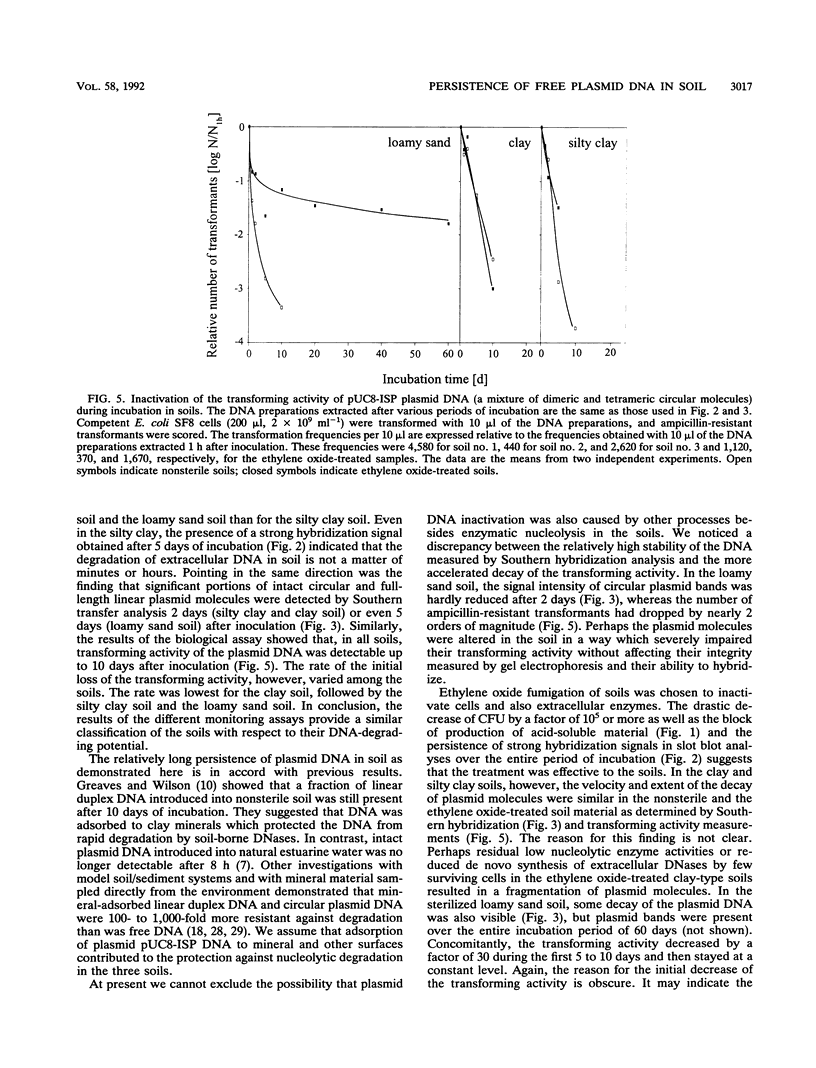
The persistence and stability of free plasmid pUC8-ISP DNA introduced into 10-g samples of various soils and kept at 23°C were monitored over a period of 60 days. The soils were sampled at a plant science farm and included a loamy sand soil (no. 1), a clay soil (no. 2), and a silty clay soil (no. 3). Four different methods allowed monitoring of (i) the production of acid-soluble radioactive material from [3H]thymidine-labeled plasmid DNA, (ii) the decrease of hybridizing nucleotide sequences in slot blot analysis, (iii) the loss of plasmid integrity measured by Southern hybridization, and (iv) the decay of the biological activity as determined by transformation of Ca2+-treated Escherichia coli cells with the DNA extracted from soil. Acid-soluble material was not produced within the first 24 h but then increased to 45% (soil no. 1), 27% (soil no. 2), and 77% (soil no. 3) until the end of incubation. A quite parallel loss of material giving a slot blot hybridization signal was observed. Southern hybridization indicated that after 1 h in the soils, plasmid DNA was mostly in the form of circular and full-length linear molecules but that, depending on the soil type, after 2 to 5 days full-length plasmid molecules were hardly detectable. The transforming activity of plasmid DNA reextracted from the soils followed inactivation curves over 2 to 4 orders of magnitude and dropped below the detection limit after 10 days. The inactivation was slower in soil no. 2 (28.2-h half-life time of the transforming activity of a plasmid molecule) than in soils no. 3 (15.1 h) and no. 1 (9.1 h). The studies provide data on the persistence of free DNA molecules in natural bacterial soil habitats. The data suggest that plasmid DNA may persist long enough to be available for uptake by competent recipient cells in situ.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CATLIN B. W. Extracellular deoxyribonucleic acid of bacteria and a deoxyribonuclease inhibitor. Science. 1956 Sep 7;124(3219):441–442. doi: 10.1126/science.124.3219.441. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb W. D., Streips U. N., Doyle R. J. Selective enrichment for genetic markers in DNA released by competent cultures of Bacillus subtilis. Mol Gen Genet. 1977 Oct 20;155(2):179–183. doi: 10.1007/BF00393157. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Lorenz M. G., Aardema B. W., Wackernagel W. Highly efficient genetic transformation of Bacillus subtilis attached to sand grains. J Gen Microbiol. 1988 Jan;134(1):107–112. doi: 10.1099/00221287-134-1-107. [DOI] [PubMed] [Google Scholar]
- Lorenz M. G., Gerjets D., Wackernagel W. Release of transforming plasmid and chromosomal DNA from two cultured soil bacteria. Arch Microbiol. 1991;156(4):319–326. doi: 10.1007/BF00263005. [DOI] [PubMed] [Google Scholar]
- Lorenz M. G., Reipschläger K., Wackernagel W. Plasmid transformation of naturally competent Acinetobacter calcoaceticus in non-sterile soil extract and groundwater. Arch Microbiol. 1992;157(4):355–360. doi: 10.1007/BF00248681. [DOI] [PubMed] [Google Scholar]
- Lorenz M. G., Wackernagel W. Adsorption of DNA to sand and variable degradation rates of adsorbed DNA. Appl Environ Microbiol. 1987 Dec;53(12):2948–2952. doi: 10.1128/aem.53.12.2948-2952.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. G., Wackernagel W. High Frequency of Natural Genetic Transformation of Pseudomonas stutzeri in Soil Extract Supplemented with a Carbon/Energy and Phosphorus Source. Appl Environ Microbiol. 1991 Apr;57(4):1246–1251. doi: 10.1128/aem.57.4.1246-1251.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. G., Wackernagel W. Natural genetic transformation of Pseudomonas stutzeri by sand-adsorbed DNA. Arch Microbiol. 1990;154(4):380–385. doi: 10.1007/BF00276535. [DOI] [PubMed] [Google Scholar]
- Paul J. H., Cazares L., Thurmond J. Amplification of the rbcL gene from dissolved and particulate DNA from aquatic environments. Appl Environ Microbiol. 1990 Jun;56(6):1963–1966. doi: 10.1128/aem.56.6.1963-1966.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., David A. W. Production of extracellular nucleic acids by genetically altered bacteria in aquatic-environment microcosms. Appl Environ Microbiol. 1989 Aug;55(8):1865–1869. doi: 10.1128/aem.55.8.1865-1869.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Frischer M. E., Thurmond J. M. Gene transfer in marine water column and sediment microcosms by natural plasmid transformation. Appl Environ Microbiol. 1991 May;57(5):1509–1515. doi: 10.1128/aem.57.5.1509-1515.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski G., Lorenz M. G., Wackernagel W. Adsorption of plasmid DNA to mineral surfaces and protection against DNase I. Appl Environ Microbiol. 1991 Apr;57(4):1057–1061. doi: 10.1128/aem.57.4.1057-1061.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayler G. S., Layton A. C. Environmental application of nucleic acid hybridization. Annu Rev Microbiol. 1990;44:625–648. doi: 10.1146/annurev.mi.44.100190.003205. [DOI] [PubMed] [Google Scholar]
- Sayler G. S., Lund L. C., Shiaris M. P., Sherrill T. W., Perkins R. E. Comparative effects of Aroclor 1254 (polychlorinated biphenyls) and phenanthrene on glucose uptake by freshwater microbial populations. Appl Environ Microbiol. 1979 May;37(5):878–885. doi: 10.1128/aem.37.5.878-885.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G. J., Sinigalliano C. D. Detection of horizontal gene transfer by natural transformation in native and introduced species of bacteria in marine and synthetic sediments. Appl Environ Microbiol. 1990 Jun;56(6):1818–1824. doi: 10.1128/aem.56.6.1818-1824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotzky G., Babich H. Survival of, and genetic transfer by, genetically engineered bacteria in natural environments. Adv Appl Microbiol. 1986;31:93–138. doi: 10.1016/s0065-2164(08)70440-4. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]




