Abstract
NY-ESO-1, a member of the cancer–testis family of antigens, is expressed in a subset of a broad range of different human tumor types. Patients with advanced NY-ESO-1-expressing tumors frequently develop humoral immunity to NY-ESO-1, and three HLA A2-restricted peptides were defined previously as targets for cytotoxic CD8+ T cells in a melanoma patient with NY-ESO-1 antibody. The objectives of the present study were (i) to develop enzyme-linked immunospot (ELISPOT) and tetramer assays to measure CD8+ T cell responses to NY-ESO-1, (ii) to determine the frequency of CD8+ T cell responses to NY-ESO-1 in a series of HLA-A2 patients with NY-ESO-1 expressing tumors, (iii) to determine the relation between CD8+ T cell and humoral immune responses to NY-ESO-1, and (iv) to compare results of NY-ESO-1 ELISPOT assays performed independently in two laboratories with T cells from the same patients. NY-ESO-1 ELISPOT and tetramer assays with excellent sensitivity, specificity, and reproducibility have been developed and found to correlate with cytotoxicity assays. CD8+ T cell responses to HLA-A2-restricted NY-ESO-1 peptides were detected in 10 of 11 patients with NY-ESO-1 antibody, but not in patients lacking antibody or in patients with NY-ESO-1-negative tumors. The results of ELISPOT assays were concordant in the two laboratories, providing the basis for standardized monitoring of T cell responses in patients receiving NY-ESO-1 vaccines.
Major progress in the identification and characterization of human tumor antigens has occurred over the past decade (1). The development of approaches to analyzing humoral (2) and cellular (3) immune reactivity to cancer in the context of the autologous host has led to the molecular characterization of tumor antigens recognized by CD8+ T cells (4) and antibody (5). As a consequence of these advances, it is now possible to classify human tumor antigens, and the majority of antigens defined to date fall into one or more of the following categories: (i) differentiation antigens, e.g., tyrosinase (6), Melan-A/MART-1 (7, 8), and gp100 (9); (ii) mutational antigens, e.g., CDK4 (10), β-catenin (11), caspase-8 (12), and p53 (13); (iii) amplification antigens, e.g., HER2/neu (14) and p53 (15); (iv) splice variant antigens (16); (v) viral antigens, e.g., HPV (17) and EBV (18); and (vi) cancer–testis (CT) antigens, e.g., MAGE (19) and NY-ESO-1 (20). The defining characteristics of CT antigens are high levels of expression in male germ cells but generally not in other normal tissues and aberrant expression in a variable proportion of a wide range of different cancer types. CT antigens first were recognized as targets for autologous cytotoxic T lymphocytes in a melanoma patient with an unusual clinical course (21). Analysis of humoral immune reactivity to human cancer by SEREX (serological screening of cDNA expression libraries) (5) also has uncovered a number of human tumor antigens with characteristics of CT antigens (22). To date, 10 genes or gene families encoding CT antigens have been recognized: MAGE (19), BAGE (23), GAGE (24), SSX (5), NY-ESO-1/LAGE-1 (20, 25), SCP-1 (26), CT7/MAGE-C1 (27, 28), CT8 (U. Sahin, personal communication), CT9 (29), and CT10/MAGE-C2 (30). Six of these CT systems are known to be coded for by genes on the X chromosome. The only CT antigen with a known function is SCP-1, a synaptonemal complex protein involved in chromosome reduction in meiosis (26).
NY-ESO-1, the focus of the present study, was identified during a SEREX analysis of an esophageal cancer (20). The gene for NY-ESO-1 maps to Xq28 (31) and codes for an 18-kDa protein (20). NY-ESO-1 mRNA expression is found in 20–30% of melanomas, lung, breast, ovarian, and bladder cancers, and other tumor types but, like other CT antigens, rarely in colon cancer or renal cancer (20). In a survey of sera from normal individuals and cancer patients, antibodies to NY-ESO-1 were found in 40–50% of patients with advanced NY-ESO-1-expressing tumors (32). One patient with high-titered NY-ESO-1 antibodies was found to have HLA-A2-restricted cytotoxic T lymphocytes against autologous NY-ESO-1-expressing melanoma cells, and three HLA-A2-restricted NY-ESO-1 peptides were identified as the target epitopes recognized by these cytotoxic T lymphocytes (33).
In the present study, we have analyzed CD8+ T cell responses to NY-ESO-1 by enzyme-linked immunospot (ELISPOT), cytotoxicity, and tetramer assays and the humoral immune responses by ELISA and Western blots. Our findings show that NY-ESO-1 elicits a strong, integrated humoral and cellular immune response in a high proportion of patients with NY-ESO-1-expressing tumors.
Materials and Methods
Tumor Typing for NY-ESO-1 mRNA.
Expression of NY-ESO-1 mRNA in tumor specimens was assessed by reverse transcription–PCR, using previously described primers (20).
Assays for NY-ESO-1 Antibody.
NY-ESO-1 serum antibodies were assayed by ELISA and Western blots, using NY-ESO-1 recombinant protein purified from Escherichia coli (32).
Peptides.
Two HLA-A2-restricted NY-ESO-1 peptides, p157–165 (SLLMWITQC) and p157–167 (SLLMWITQCFL), were selected to analyze the CD8+ T cell responses to NY-ESO-1 (34). For control purposes, the following HLA-A2-restricted peptides were included: flu matrix peptide p58–66 (GILGFVFTL), Melan-A/MART-1 peptide p26–35 (EAAGIGILTV), and MAGE-3 peptide p271–279 (FLWGPRALV). All peptides were synthesized by Multiple Peptide Systems (San Diego), with a purity of >86% as determined by reverse-phase HPLC.
Peptide Presensitization.
CD8+ T lymphocytes were separated from peripheral blood lymphocyte (PBLs) by antibody-coated magnetic beads (Minimacs; Miltenyi Biotec, Auburn, CA) and seeded into 48-well plates (Corning) at a concentration of 2.5 × 105 cells per well in RPMI medium 1640 supplemented with 10% human serum, l-asparagine (50 mg/liter), l-arginine (242 mg/liter), and l-glutamine (300 mg/liter). As antigen-presenting cells, PBLs depleted of CD8+ T cells were irradiated and incubated with 2.5 μg/ml β2-microglobulin and 10 μg/ml peptide for 1 h at room temperature and added to plates at a concentration of 1 × 106 cells per well. After 24 h, IL-2 and IL-7 (2.5 ng/ml and 10 ng/ml, respectively; Biotest Pharma, Dreieich, Germany) were added to the culture wells.
ELISPOT Assay.
Flat-bottomed, 96-well nitrocellulose plates (Millititer; Millipore) were coated with IFN-γ mAb (2 μg/ml, 1-D1K; MABTECH, Stockholm) and incubated overnight at 4°C. After washing with PBS, plates were blocked with 10% human AB serum for 1 h at 37°C. Fifty thousand CD8+ cells, either presensitized with peptide for 6 days (see above) or not presensitized, and 5 × 104 T2 cells pulsed with 10 μg/ml peptide were added to each well and incubated for 20 h in RPMI medium 1640 lacking IL-2 and human serum. After incubation, the plates were washed thoroughly with PBS to remove cells, and IFN-γ mAb (0.2 μg/ml, 7-B6-1-biotin; MABTECH) was added to each well. After incubation for 2 h at 37°C, plates were washed and developed with streptavidin-alkaline phosphatase (1 μg/ml; MABTECH) for 1 h at room temperature. After washing, substrate (5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium; Sigma) was added and incubated for 5 min. After washing, the dark-violet spots were counted under the microscope.
Cytotoxicity Assay.
For assessment of cytotoxicity, CD8+ T cells presensitized with peptide (see above) were restimulated on day 7 with 2 μg/ml peptide and cultured for a further 5 days in RPMI medium 1640 with additional IL-2 (5 ng/ml). On day 12, presensitized or nonsensitized CD8+ T cells were incubated with 1 × 106 T2 cells, pulsed with 10 μg/ml peptide, and labeled with 100 μCi of Na 51CrO4 (DuPont) for 4 h with 5% CO2 at 37°C. Unlabeled K562 cells were added at a ratio of 40:1 to block NK activity. Nonpeptide-pulsed T2 cells also were used as control targets. The percentage of specific 51Cr release was determined by the following formula: percent specific 51Cr release = (experimental 51Cr release − spontaneous 51Cr release) × 100/(maximum 51Cr release − spontaneous 51Cr release). Maximum 51Cr release was obtained by adding 100 μl of 1% Nonidet P-40 (Sigma) to labeled target cells. Spontaneous 51Cr release ranged from 3% to 10%.
Tetramer Synthesis.
HLA-A2 tetrameric complexes were synthesized as described previously (34). Briefly, purified HLA heavy chain and β2-microglobulin were synthesized in a prokaryotic expression system (pHN1). The heavy chain was modified by deletion of the transmembrane cytosolic tail and COOH-terminal addition of a sequence containing the BirA enzymatic biotinylation site. Heavy chain, β2-microglobulin, and peptide were refolded by dilution and biotinylated by BirA (Avidity, Denver) in the presence of biotin, adenosine 5′-triphosphate. The 45-kDa refolded product was isolated by size-exclusion chromatography. Extravidin-phycoerythrin conjugate (Sigma) was added at a 1:4 molar ratio. Tetramers were assembled with NY-ESO-1-derived peptide p157–165, Melan-A/MART-1 peptide p26–35, and MAGE-3 peptide p271–279.
Tetramer Assay.
CD8+ T cells were presensitized with peptide as described above. The cultures were refed with 2.5 μg/ml IL-2 and 10 ng/ml IL-7 on days 4 and 7, and the presensitization period was extended to 10 days. Sensitized (5 × 104 cells per sample) and nonsensitized (1 × 106 cells per sample) PBLs were stained with phycoerythrin-labeled tetramer for 15 min at 37°C before addition of Tricolor-CD8 mAb (Caltag, South San Francisco, CA) for 15 min on ice. After washing, stained cells were analyzed by flow cytometry (FACSCalibur; Becton Dickinson).
Results
Typing for NY-ESO-1 Expression and NY-ESO-1 Antibody.
Thirty-six HLA-A2-positive patients with stage IV melanoma or other types of advanced cancer were typed for NY-ESO-1 mRNA expression by reverse transcription–PCR and for NY-ESO-1 antibody. Tumors from 27 of 36 patients expressed NY-ESO-1 mRNA, and 11 of these patients had NY-ESO-1 antibody. NY-ESO-1 antibody was not detected in patients with NY-ESO-1-negative tumors. These findings confirm and extend the results of previous serological surveys for NY-ESO-1 antibody (32).
NY-ESO-1 ELISPOT Assay.
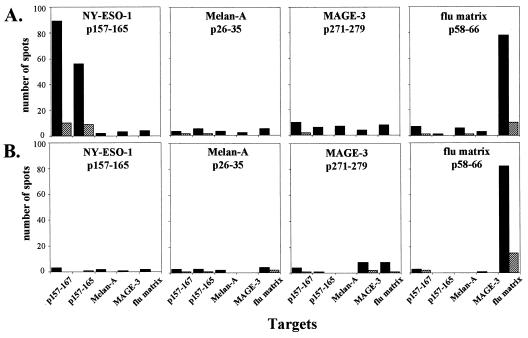
A range of variables involved in the ELISPOT assay initially was explored to define optimal conditions for measuring CD8+ T cell responses to NY-ESO-1. An assay has been developed that shows requisite sensitivity and specificity and a high degree of reproducibility. High background (e.g., spots in the absence of specific antigen stimulation) has been a general problem with ELISPOT, and this has been dealt with in past studies by subtracting background reactivity from reactivity after specific stimulation. In the ELISPOT assay we have developed for NY-ESO-1, the background is extremely low and no subtraction is required. Fig. 1 illustrates ELISPOT assays for NY-ESO-1 reactivity with CD8+ T cells from two stage IV melanoma patients. Melanoma specimens from both patients typed NY-ESO-1 mRNA-positive, but NY-ESO-1 antibody was detected in only one of them. CD8+ and CD8− cell populations were prepared from PBLs, and the purified CD8+ T cells were presensitized with autologous irradiated CD8− cells pulsed with HLA-A2-restricted peptides derived from NY-ESO-1, Melan-A/MART-1, MAGE-3, or flu matrix. The flu matrix peptide served as a specificity control for patients with preexisting flu reactivity, providing assurance that the culture and assay conditions were permissive. After 6 days of presensitization, CD8+ T cells were recovered and mixed with T2 cells pulsed with the series of peptides and tested for IFN-γ release in ELISPOT assays. Specificity was assessed by comparing CD8+ T cell reactivity against the presensitizing peptide to reactivity against unrelated peptides. As shown in Fig. 1, the patient with NY-ESO-1 antibody, NW731, had strong CD8+ T cell reactivity to T2 cells pulsed with NY-ESO-1 p157–167 or p157–165, but not with Melan-A/MART-1 or MAGE-3 peptides. The extent of the NY-ESO-1 response was comparable to that against the flu matrix peptide. In contrast, the patient without NY-ESO-1 antibody, NW681, had no demonstrable reactivity against NY-ESO-1 peptides, but did show a strong response to the flu matrix peptide.
Figure 1.
CD8+ T cell reactivity to HLA-A2-restricted peptides in ELISPOT IFN-γ assays. (A) CD8+ T cells from NW731, a patient with NY-ESO-1-positive melanoma and NY-ESO-1 antibody. (B) CD8+ T cells from NW681, a patient with NY-ESO-1-positive melanoma and no NY-ESO-1 antibody. CD8+ T cells were presensitized with NY-ESO-1 p157–165, Melan-A/MART-1 p26–35, MAGE-3 p271–279, or flu matrix p58–66 and tested against T2 cells pulsed with the peptide panel shown at the bottom of the figure (including NY-ESO-1 p157–167). Effector-to-target cell ratios in these assays were 1:1 (solid bars) and 0.2:1 (hatched bars).
Influence of Peptide Presensitization on NY-ESO-1 Reactivity.
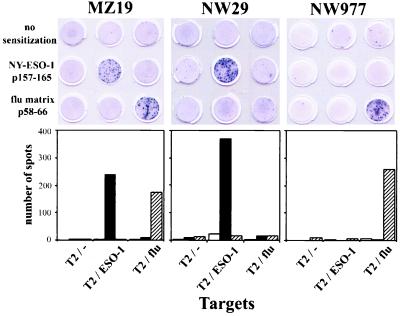
Fig. 2 illustrates the importance of presensitization with peptides for demonstrating NY-ESO-1 reactivity. CD8+ T cells from three patients with NY-ESO-1-positive tumors and antibody were tested for NY-ESO-1 reactivity, either directly ex vivo or after presensitization with NY-ESO-1 or flu matrix peptides. No or minimal CD8+ T cell reactivity was observed in the absence of peptide presensitization. In contrast, strong NY-ESO-1 reactivity was seen in two of three patients after presensitization of CD8+ T cells with NY-ESO-1 peptides. Similarly, the response against flu matrix peptide was augmented greatly by specific peptide presensitization. Thus, although low levels of reactivity have been seen with CD8+ T cells from a few patients, the presensitization step with NY-ESO-1 peptides appeared essential to optimize the assay.
Figure 2.
Effect of presensitization of CD8+ T cells with HLA-A2-restricted NY-ESO-1 p157–165 and flu matrix p58–66 on ELISPOT assays. Results with CD8+ T cells from three patients with NY-ESO-1 expressing stage IV melanoma and NY-ESO-1 antibody are shown. Fifty thousand CD8+ T cells, with or without presensitization with NY-ESO-1 p157–165 or flu matrix p58–66, were cocultured with 50,000 T2 cells alone or pulsed with NY-ESO-1 p157–165 or flu matrix p58–66. (Upper) Photographs of wells in an ELISPOT assay. (Lower) Histograms with no presensitization (open bars), presensitization with NY-ESO-1 p157–165 (solid bar), or presensitization with flu matrix p58–66 (hatched bar). Each bar represents average of spots in duplicate wells.
Frequency of CD8+ T Cell Reactivity Against NY-ESO-1.
Table 1 summarizes the results of ELISPOT assays with CD8+ T cells from 27 patients with NY-ESO-1-positive tumors and 9 patients with NY-ESO-1-negative tumors. No ELISPOT reactivity was detected in patients with NY-ESO-1-negative tumors. Ten of the 27 patients with NY-ESO-1-positive tumors showed strong ELISPOT reactivity, and all 10 patients with ELISPOT reactivity had NY-ESO-1 antibody. There was only one patient (NW977) with a discrepancy between antibody status (positive) and ELISPOT (negative) reactivity. As yet, the contrary has not been observed, e.g., antibody-negative/ELISPOT-positive.
Table 1.
CD8+ T cell reactivity to HLA-A2-restricted NY-ESO-1 p157-165 in ELISPOT assays
| Patient* | Peptide presensitization†
|
Cytotoxicity‡ | |
|---|---|---|---|
| − | + | ||
| NY-ESO-1 mRNA-positive/Ab-positive | |||
| MZ19 | 0 (0) | 239 (2) | + |
| NW14 | ND§ | 240 (1) | + |
| NW29 | 9 (5) | 367 (22) | + |
| NW37 | 15 (2) | 60 (3) | + |
| NW516 | ND | 540 (10) | + |
| NW519 | 6 (0) | 285 (8) | + |
| NW731 | ND | 69 (1) | + |
| NW743 | ND | 55 (5) | + |
| NW889 | 37 (9) | 55 (12) | + |
| NW903 | 0 (0) | 700 (12) | + |
| NW977 | 0 (0) | 0 (0) | − |
| NY-ESO-1 mRNA-positive/Ab-negative | |||
| NW28 | 0 (0) | 0 (1) | − |
| NW33 | 6 (10) | 0 (0) | − |
| NW46 | ND | 0 (0) | − |
| NW208 | 4 (20) | 0 (0) | ND |
| NW235 | 4 (8) | 0 (0) | − |
| NW241 | 0 (0) | 15 (8) | − |
| NW315 | 0 (0) | 5 (0) | − |
| NW388 | ND | 4 (5) | ND |
| NW415 | 2 (0) | 0 (0) | − |
| NW604 | 12 (16) | 0 (1) | − |
| NW681 | 24 (20) | 0 (0) | − |
| NW726 | 0 (0) | 0 (0) | − |
| NW745 | 0 (0) | 0 (0) | − |
| NW789 | 1 (0) | 0 (0) | − |
| NW836 | 0 (0) | 0 (0) | − |
| NW989 | 0 (0) | 0 (0) | − |
| NY-ESO-1 mRNA-negative/Ab-negative | |||
| NW30 | 0 (0) | 0 (0) | − |
| NW44 | 0 (0) | 0 (0) | − |
| NW45 | 0 (0) | 0 (0) | − |
| NW50 | ND | 0 (0) | − |
| NW51 | ND | 0 (0) | − |
| NW145 | 0 (0) | 0 (0) | ND |
| NW309 | ND | 0 (0) | ND |
| NW379 | ND | 0 (0) | ND |
| NW449 | 0 (0) | 0 (0) | ND |
ND, not determined.
All patients with stage IV melanoma, except for NW516 (breast cancer), NW519 (non-small-cell lung cancer), NW889 and NW989 (ovarian cancer), and NW977 and NW315 (prostatic cancer).
† 50,000 CD8+ T cells with (+) or without (−) presensitization with NY-ESO-1 p157-165 were tested for IFN-γ release after 20 h of incubation with NY-ESO-1 p157-165 pulsed T2 cells. The results shown here are an average of duplicate wells. Values in parentheses indicate average number of spots with nonpeptide pulsed T2 cells. Assays were repeated at least twice, with similar results.
‡ Cytotoxicity of CD8+ T cell presensitized with NY-ESO-1 p157-165 against T2 cells pulsed with NY-ESO-1 p157-165. +, Greater than 20% specific lysis against NY-ESO-1 p157-165 pulsed T2 target cells. Presensitized CD8+ T cells from patients NW14, NW29, NW731, and NW37 showed specific cytotoxicity for NY-ESO-1-positive melanoma cell lines (NW-MEL-38 or SK-MEL-37).
To determine the reproducibility of the NY-ESO-1 ELISPOT assay, PBLs from 21 patients in this series were assayed in two separate laboratories (Frankfurt and New York). The results were concordant, both in terms of presence or absence of ELISPOT reactivity and the magnitude of the response.
NY-ESO-1 Cytotoxicity Tests.
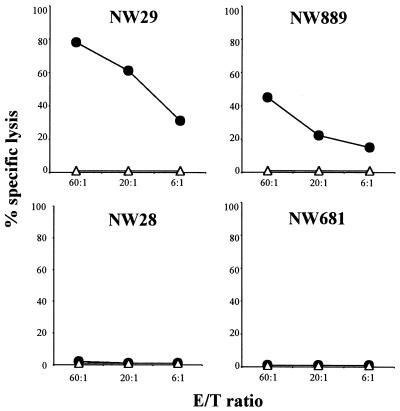
Fig. 3 shows cytotoxicity tests with CD8+ T cells from four patients with NY-ESO-1+ tumors. Two of the patients, NW29 and NW889, had NY-ESO-1 antibody, and two patients, NW28 and NW681, had no antibody. CD8+ T cells were presensitized with NY-ESO-1 p157–165 for 12 days and tested for cytotoxic activity against T2 target pulsed with the NY-ESO-1 p157–165. NY-ESO-1-specific cytotoxicity was observed in the two patients with NY-ESO-1 antibody, but not in the two patients lacking NY-ESO-1 antibody. Table 1 summarizes the results of cytotoxic tests with 30 patients in this series. No NY-ESO-1 cytotoxicity was found with CD8+ T cells from patients with NY-ESO-1-negative tumors or from patients with NY-ESO-1-positive tumors but lacking NY-ESO-1 antibody. There was a complete concordance between ELISPOT and cytotoxicity assays.
Figure 3.
Cytotoxicity tests with CD8+ T cells from four HLA-A2 patients with NY-ESO-1-positive melanomas. Patients NW29 and NW889 had NY-ESO-1 antibody, and patients NW28 and NW681 did not. Cytotoxicity was measured by standard 51Cr release assay, and values represent averages of duplicate wells (▵, T2 cells; ●, T2 cells pulsed with NY-ESO-1 p157–165).
NY-ESO-1 Tetramer Assay.
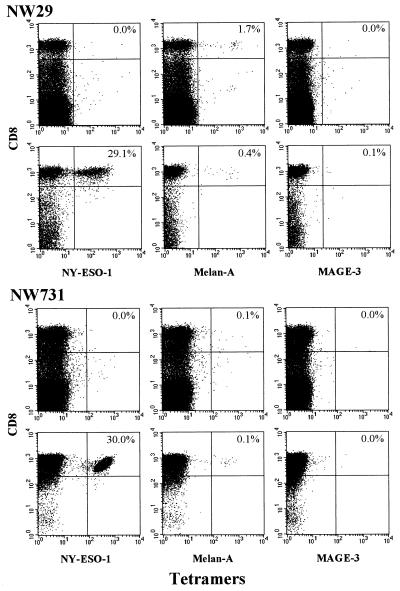
HLA-A2 tetramers were prepared with the following HLA-A2-restricted peptides: NY-ESO-1 p157–165, Melan-A/MART-1 p26–35, and MAGE-3 p271–279. Fig. 4 illustrates tests with CD8+ T cells from two melanoma patients with NY-ESO-1-positive tumors and NY-ESO-1 antibody. Before peptide presensitization, no NY-ESO-1 tetramer binding could be observed. However, after presensitization with NY-ESO-1 peptides, remarkably high levels of specific NY-ESO-1 tetramer binding were found. Table 2 summarizes the results of tetramer, ELISPOT, and cytotoxicity assays on 18 patients in this series. NY-ESO-1 tetramers did not bind to CD8+ T cells from patients without NY-ESO-1 antibody. In patients with NY-ESO-1 antibody, NY-ESO-1 tetramer binding was detected in four of the six patients tested. NY-ESO-1 tetramer-positive patients also were reactive in ELISPOT and cytotoxicity assays.
Figure 4.
Tetramer analysis of CD8+ T cells from two stage IV melanoma patients, NW29 and NW731, with NY-ESO-1-positive tumors and NY-ESO-1 antibody. Tests with unsensitized PBLs (Upper) and CD8+ T cells presensitized with NY-ESO-1 p157–165 (Lower) for each patient are shown. Samples were double-stained with phycoerythrin-labeled tetramers and anti-Tricolor-CD8 mAb. Tetramers were prepared with NY-ESO-1 p157–165, Melan-A p26–35, and MAGE-3 p271–279 as indicated at the bottom of each grid. Values indicate percentage of tetramer-positive CD8+ T cells.
Table 2.
CD8+ T cell reactivity to NY-ESO-1 HLA-A2-restricted peptides: Summary of tetramer, ELISPOT, and cytotoxicity assays
| Patient | Tetramer* | ELISPOT* | Cytotoxicity |
|---|---|---|---|
| NY-ESO-1 mRNA-positive/Ab-positive | |||
| NW14 | + | + | + |
| NW29 | + | + | + |
| NW516 | + | + | + |
| NW731 | + | + | + |
| NW37 | − | + | + |
| NW977 | − | − | − |
| NY-ESO-1 mRNA-positive/Ab-negative | |||
| NW28 | − | − | − |
| NW33 | − | − | − |
| NW46 | − | − | − |
| NW315 | − | − | − |
| NW415 | − | − | − |
| NW681 | − | − | − |
| NW726 | − | − | − |
| NY-ESO-1 mRNA-negative/Ab-negative | |||
| NW30 | − | − | − |
| NW44 | − | − | − |
| NW45 | − | − | − |
| NW50 | − | − | − |
| NW51 | − | − | − |
Discussion
With the identification of a growing number of human tumor antigens, the challenge is to develop vaccination strategies that induce protective or therapeutic immunity. To do so, standardized methods of monitoring humoral and cellular immune responses to tumor antigens are essential, and considerable progress has been made toward developing reliable assays to analyze antibody and CD8+ T cell responses. Until recently, cytotoxicity assays in conjunction with limiting dilution analysis represented the primary method to measure specific CD8+ T cell responses. Now, cytokine release assays and tetramer analysis provide powerful new ways to assess the specificity and magnitude of CD8+ T cell as well as CD4+ T cell responses. Thus, with current technologies it is now possible to characterize distinct aspects of the T cell response to antigens. HLA/peptide tetramers are able to detect the presence of specific T cell receptors on the cell surface, cytokine release assays reflect the capacity of T cells to be activated in response to antigen, and cytotoxicity tests indicate that the specifically activated T cell has the machinery to kill target cells. Together with the monitoring of humoral immunity, these approaches in conjunction with phenotyping T cells for other markers of T cell development and function allow tumor immunologists to obtain a comprehensive picture of the integrated immune response of humans to human tumor antigens.
In terms of eliciting humoral and cellular immune responses, NY-ESO-1 appears to be the most immunogenic CT antigen and one of the most immunogenic human tumor antigens defined to date. In our past study of humoral immunity to NY-ESO-1, 40–50% of patients with advanced NY-ESO-1-expressing tumors were found to have NY-ESO-1 antibody. In the present study, we find that a high percentage of patients with NY-ESO-1 antibody also have detectable CD8+ T cell responses to known HLA-A2-restricted NY-ESO-1 peptides. The picture that emerges for NY-ESO-1 immune reactivity can be summarized as follows. (i) Antibody and CD8+ T cell responses to NY-ESO-1 occur only in patients with NY-ESO-1-expressing tumors. (ii) CD8+ T cell responses to NY-ESO-1 have not been detected in patients without NY-ESO-1 antibody. (iii) Humoral immunity to NY-ESO-1 in the absence of CD8+ T cells to known HLA-A2-restricted NY-ESO-1 peptides has been observed in only one patient. In this patient, a NY-ESO-1-specific CD8+ T cell response may be occurring but possibly directed toward other NY-ESO-1-derived peptides. (iv) There is a strong concordance in the results of ELISPOT, cytotoxicity, and tetramer assays for HLA-A2-restricted CD8+ T cell responses to NY-ESO-1. (v) CD8+ T cells with a NY-ESO-1 tetramer-positive, ELISPOT-negative, cytotoxicity-negative phenotype have not been detected, suggesting that T cell anergy to NY-ESO-1 is uncommon.
The next step in monitoring CD8+ T cell response to NY-ESO-1 is the definition of additional HLA-restricted peptide targets. What is needed is a general method for peptide identification that can be used in patients with any HLA haplotype, and we are evaluating an approach that involves transfection of autologous antigen-presenting cells with NY-ESO-1 coding vectors. Because an antibody response to NY-ESO-1 correlates with a CD8+ T cell response to NY-ESO-1 in HLA-A2-positive patients, the presence of NY-ESO-1 antibody can be used to identify non-HLA-A2 patients with a likely CD8+ T cell response. Wang et al. (35) have identified an HLA-A31-restricted NY-ESO-1 peptide coded for by an alternative reading frame. To determine whether a NY-ESO-1 product derived from this alternative reading frame might elicit antibody and thus explain the apparent seronegativity of certain patients with NY-ESO-1-expressing tumors, we have prepared a NY-ESO-1 fusion protein derived from this alternative reading frame. No antibodies have been detected against this product.
With the development of secure immunological monitoring methodologies for NY-ESO-1, we can begin to address some key issues related to the causes and consequences of the strong immunogenicity of NY-ESO-1. Previous studies have shown that patients with NY-ESO-1 antibody tend to have advanced-stage cancer (32, 36) and that NY-ESO-1 antibody titers fall with removal or successful therapy of NY-ESO-1-expressing tumors (36). This relation of seropositivity and tumor burden suggests that NY-ESO-1 is highly dependent on persistent antigen stimulation. In immunohistochemical analysis, NY-ESO-1 expression, like the expression of another CT antigen, MAGE, tends to be highly heterogeneous, with some tumors showing antigen expression in only a small population of tumor cells (37). A study is ongoing to determine whether there is any correlation between the pattern of NY-ESO-1 antigen expression in tumors and the presence/strength or absence of NY-ESO-1 immunity. Finally, what influence does NY-ESO-1 have on the clinical course of patients with NY-ESO-1-expressing tumors? Because of the variable behavior of cancer in individual patients and the unpredictable influence of therapeutic interventions, a definite answer to this question will come only after a detailed survey of larger numbers of patients. One possibility we will be looking for is evidence that NY-ESO-1 immunity provides a strong, selective pressure for the emergence of NY-ESO-1/HLA-A2 antigen loss variants, comparable to what has been seen in patients vaccinated with melanocyte differentiation antigens (38).
Abbreviations
- ELISPOT
enzyme-linked immunospot
- PBL
peripheral blood lymphocyte
- SEREX
serological screening of cDNA expression libraries
- CT
cancer–testis
References
- 1.Boon T, Old L J. Curr Opin Immunol. 1997;9:681–683. doi: 10.1016/s0952-7915(97)80049-0. [DOI] [PubMed] [Google Scholar]
- 2.Carey T E, Takahashi T, Resnick L A, Oettgen H F, Old L J. Proc Natl Acad Sci USA. 1976;73:3278–3282. doi: 10.1073/pnas.73.9.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knuth A, Danowski B, Oettgen H F, Old L J. Proc Natl Acad Sci USA. 1984;81:3511–3515. doi: 10.1073/pnas.81.11.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 5.Sahin U, Türeci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Proc Natl Acad Sci USA. 1995;92:11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brichard V, Van Pel A, Wölfel T, Wölfel C, De Plaen E, Lethe B, Coulie P, Boon T. J Exp Med. 1993;178:489–495. doi: 10.1084/jem.178.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coulie P G, Brichard V, Van Pel A, Wölfel T, Schneider J, Traversari C, Mattei S, De Plaen E, Lurquin C, Szikora J P, et al. J Exp Med. 1994;180:35–42. doi: 10.1084/jem.180.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawakami Y, Eliyahu S, Delgado C H, Robbins P F, Rivoltini L, Topalian S L, Miki T, Rosenberg S A. Proc Natl Acad Sci USA. 1994;91:3515–3519. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawakami Y, Eliyahu S, Jennings C, Sakaguchi K, Kang X, Southwood S, Robbins P F, Sette A, Appella E, Rosenberg S A. J Immunol. 1995;154:3961–3968. [PubMed] [Google Scholar]
- 10.Wölfel T, Hauer M, Schneider J, Serrano M, Wölfel C, Klehmann-Hieb E, De Plaen E, Hankeln T, Meyer zum Büschenfelde K H, Beach D. Science. 1995;269:1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 11.Robbins P F, El-Gamil M, Li Y F, Kawakami Y, Loftus D, Appella E, Rosenberg S A. J Exp Med. 1996;183:1185–1192. doi: 10.1084/jem.183.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandruzzato S, Brasseur F, Andry G, Boon T, van der Bruggen P. J Exp Med. 1997;186:785–793. doi: 10.1084/jem.186.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scanlan M J, Chen Y T, Williamson B, Güre A O, Stockert E, Gordan J D, Türeci O, Sahin U, Pfreundschuh M, Old L J. Int J Cancer. 1998;76:652–658. doi: 10.1002/(sici)1097-0215(19980529)76:5<652::aid-ijc7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 14.Cheever M A, Disis M L, Bernhard H, Gralow J R, Hand S L, Huseby E S, Qin H L, Takahashi M, Chen W. Immunol Rev. 1995;145:33–59. doi: 10.1111/j.1600-065x.1995.tb00076.x. [DOI] [PubMed] [Google Scholar]
- 15.Gnjatic S, Cai Z, Viguier M, Chouaib S, Guillet J G, Choppin J. J Immunol. 1998;160:328–333. [PubMed] [Google Scholar]
- 16.Scanlan M J, Williamson B, Jungbluth A, Stockert E, Arden K C, Viars C S, Güre A O, Gordan J D, Chen Y T, Old L J. Biochim Biophys Acta. 1999;1445:39–52. doi: 10.1016/s0167-4781(99)00033-0. [DOI] [PubMed] [Google Scholar]
- 17.Tindle R W. Curr Opin Immunol. 1996;8:643–650. doi: 10.1016/s0952-7915(96)80080-x. [DOI] [PubMed] [Google Scholar]
- 18.Lennette E T, Winberg G, Yadav M, Enblad G, Klein G. Eur J Cancer. 1995;31A:1875–1878. doi: 10.1016/0959-8049(95)00354-l. [DOI] [PubMed] [Google Scholar]
- 19.Boon T, van der Bruggen P. J Exp Med. 1996;183:725–729. doi: 10.1084/jem.183.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y T, Scanlan M J, Sahin U, Türeci O, Gure A O, Tsang S, Williamson B, Stockert E, Pfreundschuh M, Old L J. Proc Natl Acad Sci USA. 1997;94:1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herin M, Lemoine C, Weynants P, Vessiere F, Van Pel A, Knuth A, Devos R, Boon T. Int J Cancer. 1987;39:390–396. doi: 10.1002/ijc.2910390320. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y T, Scanlan M J, Obata Y, Old L J. In: Cancer Vaccines: Cancer Antigens; Identification of Human Tumor Antigens by Serological Expression Cloning (SEREX) Devita V T, Hellman S, Rosenberg S A, editors. Philadelphia: Lippincott; 1999. pp. 557–570. [Google Scholar]
- 23.Boël P, Wildmann C, Sensi M L, Brasseur R, Renauld J C, Coulie P, Boon T, van der Bruggen P. Immunity. 1995;2:167–175. doi: 10.1016/s1074-7613(95)80053-0. [DOI] [PubMed] [Google Scholar]
- 24.Van den Eynde B, Peeters O, De Backer O, Gaugler B, Lucas S, Boon T. J Exp Med. 1995;182:689–698. doi: 10.1084/jem.182.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lethé B, Lucas S, Michaux L, De Smet C, Godelaine D, Serrano A, De Plaen E, Boon T. Int J Cancer. 1998;76:903–908. doi: 10.1002/(sici)1097-0215(19980610)76:6<903::aid-ijc22>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 26.Türeci O, Sahin U, Zwick C, Koslowski M, Seitz G, Pfreundschuh M. Proc Natl Acad Sci USA. 1998;95:5211–5216. doi: 10.1073/pnas.95.9.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y T, Güre A O, Tsang S, Stockert E, Jäger E, Knuth A, Old L J. Proc Natl Acad Sci USA. 1998;95:6919–6923. doi: 10.1073/pnas.95.12.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucas S, De Smet C, Arden K C, Viars C S, Lethé B, Lurquin C, Boon T. Cancer Res. 1998;58:743–752. [PubMed] [Google Scholar]
- 29.Scanlan M J, Altorki N K, Güre A O, Williamson B, Jungbluth A J, Chen Y-T, Old L J. Cancer Lett. 2000;150:155–164. doi: 10.1016/s0304-3835(99)00385-7. [DOI] [PubMed] [Google Scholar]
- 30.Güre A O, Stockert E, Arden K C, Boyer A D, Viars C S, Scanlan M J, Old L J, Chen Y T. Int J Cancer. 2000;85:726–732. doi: 10.1002/(sici)1097-0215(20000301)85:5<726::aid-ijc21>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y T, Boyer A D, Viars C S, Tsang S, Old L J, Arden K C. Cytogenet Cell Genet. 1997;79:237–240. doi: 10.1159/000134734. [DOI] [PubMed] [Google Scholar]
- 32.Stockert E, Jäger E, Chen Y T, Scanlan M J, Gout I, Karbach J, Arand M, Knuth A, Old L J. J Exp Med. 1998;187:1349–1354. doi: 10.1084/jem.187.8.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jäger E, Chen Y T, Drijfhout J W, Karbach J, Ringhoffer M, Jäger D, Arand M, Wada H, Noguchi Y, Stockert E, et al. J Exp Med. 1998;187:265–270. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 35.Wang R F, Johnston S L, Zeng G, Topalian S L, Schwartzentruber D J, Rosenberg S A. J Immunol. 1998;161:3598–3606. [PubMed] [Google Scholar]
- 36.Jäger E, Stockert E, Zidianakis Z, Chen Y T, Karbach J, Jäger D, Arand M, Ritter G, Old L J, Knuth A. Int J Cancer. 1999;84:506–510. doi: 10.1002/(sici)1097-0215(19991022)84:5<506::aid-ijc10>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 37.Jungbluth A A, Busam K J, Kolb D, Iversen K, Coplan K, Chen Y T, Spagnoli G C, Old L J. Int J Cancer. 2000;85:460–465. [PubMed] [Google Scholar]
- 38.Jäger E, Ringhoffer M, Altmannsberger M, Arand M, Karbach J, Jäger D, Oesch F, Knuth A. Int J Cancer. 1997;71:142–147. doi: 10.1002/(sici)1097-0215(19970410)71:2<142::aid-ijc3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]