Abstract
Lsh (Hells) is closely related to SNF2/helicase family members that remodel chromatin and thus regulate gene transcription. In the adult mouse Lsh is expressed primarily in lymphoid tissue, showing the highest level in thymocytes. Lsh gene expression can be induced in thymic pro-T cells by pre-T cell receptor crosslinking and in mature T cells by T cell receptor crosslinking together with costimulation via CD28. The time course of Lsh gene and protein expression correlated closely with the onset of S phase of the cell cycle. To explore the function of Lsh during lymphoid development or activation, we deleted the Lsh gene by homologous recombination in ES cells. Fetal liver cells from Lsh−/− were used as a source of hematopoietic precursors to reconstitute lymphoid development in Rag2−/− mice. Lsh−/− (compared to Lsh+/+ or ±) chimeras showed a modest reduction in thymocyte numbers due to a partial arrest at the transition from the CD4−CD8− stage to the CD4+CD8+ stage of T cell development. Mature peripheral lymphocytes were reduced in number to ≈60% for T cells and 40% for B cells; however, V(D)J recombination of the immune receptor genes was normal. Although polyclonal activation of Lsh−/− T cells induced normal levels of cytokines, cell proliferation was severely suppressed and cells underwent apoptosis. Several genes involved in the regulation of apoptosis were expressed normally with the exception of Bcl-2 that was actually elevated. These findings demonstrate that Lsh is not obligatory for normal lymphoid development but is essential for normal proliferation of peripheral T lymphocytes.
Lymphoid-specific helicase (Lsh) is a member of the SNF2 subfamily of helicases with highest homology to the CHD, SNF2, and SNF2L-like groups within this family that are thought to act as transciptional regulators (1). Members of these LSH-related subgroups such as SNF2, Brahma and Brg-1, imitation mating-type switching (ISWI), and chromodomain-helicase-DNA binding protein (CHD) proteins function as components of large protein complexes known as SNF/SWI, NURF, or NURD complexes that disrupt mononucleosomal structures in vitro, an activity thought to relate to their chromatin-remodeling properties in vitro (2). Thus, SNF2 homologs are a major component in these complexes responsible for their ability to alter chromatin structure and thus transcription. We recently cloned Lsh/Hells from thymocytes undergoing rearrangement, using a PCR-based strategy exploiting the highly conserved helicase domains [refs. 3 and 4; note that the genetic sequence has been entered in Genbank as Lsh (lymphoid specific helicase) and in the mouse genome database as Hells (helicase, lymphoid-specific)]. Lsh mRNA was found to be preferentially expressed in lymphoid tissue in the adult mouse, which contrasts with the ubiquitous expression of many other mammalian SNF2/helicase homologs (3, 4). Lsh levels were high in thymus, the organ of T cell development during which precursor cells proceed from the CD4−CD8− stage to become mature CD4+ and CD8+ T cells after undergoing V(D)J recombination, extensive expansion, and negative and positive selectional processes (5). We hypothesized that Lsh could play a distinct role in lymphocyte development and activation by altering the chromatin structure in lymphoid cells and thus controlling T cell-specific gene expression. In this study, we examined the expression pattern of Lsh mRNA and protein during lymphoid differentiation and activation. To test the role of Lsh in lymphoid development, we deleted the Lsh gene in mice. Because Lsh−/− mice died perinatally, lymphoid development and function was studied using Lsh−/− fetal liver to reconstitute lymphoid development in Rag2−/− recipients. These studies revealed that Lsh is required for proliferation of peripheral T lymphocytes.
Methods
Mice and Cell Lines.
Generation of Lsh−/− mice and genotyping are described elsewhere. Lsh± mice housed in a pathogen-free environment were mated overnight and checked for plugs the following day (designated day 1). Animals were sacrificed at day 14–18 of gestation. For generation of Lsh−/−Rag2−/− chimeras, fetal liver cell suspensions (2.5–5 × 106 cells) from Lsh−/− embryos or littermates with ± or +/+ genotype were i.v. injected into irradiated Rag2−/− recipients (650–850 rad). Lymphoid reconstitution was analyzed in thymus, spleen, lymph nodes, and blood 4–5 wk later. SCID and Rag-2−/− mice (Jackson Laboratory) (4–8 wk of age) were injected i.v. with 200 μl of a 1:10 dilution of the 2C11 anti-CD3 antiserum (kind gift of S. K. Durum, NCI) and the thymus analyzed 6 d later. For analysis of Lsh expression in lymphoid subpopulations, fetal or adult mice of the C57BL/6J strain were used (Jackson Laboratory) (Figs. 1 and 2). EL4 (American Type Culture Collection) is a T cell line and 38B9 (kind gift by F. W. Alt, Harvard Medical School, Boston) is a pre-B cell line.
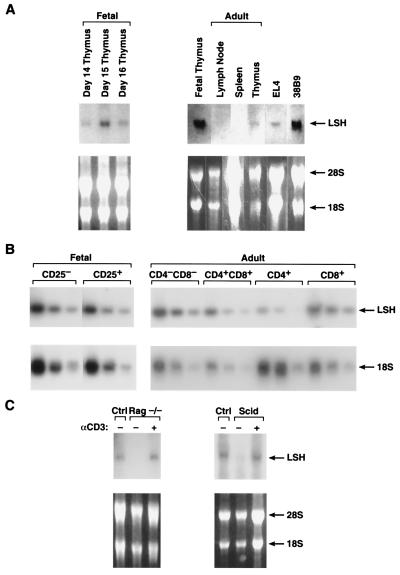
Figure 1.
Expression of Lsh mRNA in lymphoid tissue. (A) Lsh mRNA expression in fetal vs. adult lymphoid tissue. Embryonal thymi were removed at day 14, 15, or 16 of gestation (left) or thymus, spleen, and lymph node were removed from 4–8 wk old adults (right) and compared to fetal thymus (day 15). EL4 is a T cell line and 38B9 is a pre-B cell line. Total RNA was extracted and subjected to Northern analysis by using a 2.2-kb Lsh cDNA probe comprising the whole ORF. (B) Lsh mRNA expression in T cell precursor populations. Fetal (day 14) or adult thymus were stained with the indicated antibodies and sorted by flow cytometry. Total RNA was extracted and subjected to RT-PCR analysis (1/5 serial dilutions) for specific detection of Lsh or 18s (control) mRNA. (C) Induction of Lsh mRNA by pre-TCR signaling. Rag2−/− and SCID mice were injected with anti-CD3 antibodies and the thymus removed 6 d later for Northern analysis as in A.
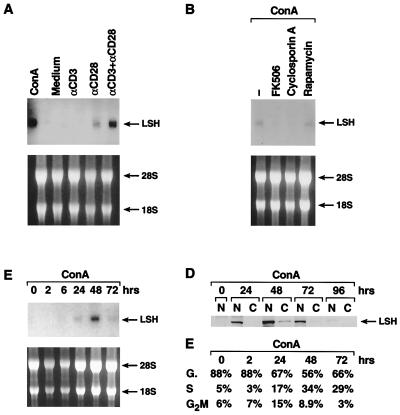
Figure 2.
Induction of LSH gene expression in mature T lymphocytes. (A) Induction of Lsh mRNA by CD3 and CD28 costimulation. Splenocytes were cultured for 48 h in the presence of the indicated stimuli and Northern analysis performed as in Fig. 1A. (B) Inhibition of Lsh mRNA by immunosuppressive drugs. Splenocytes from animals were cultured for 48 h with ConA in the presence or absence of the immunosuppressive drugs cyclosporine A, FK506, or rapamycin, and Northern analysis was performed as in Fig. 1A. (C) Time kinetics of Lsh mRNA expression. Splenocytes were stimulated for 2–72 h with ConA, and Northern analysis was performed as in Fig. 1A. (D) Time kinetics of Lsh protein expression. Splenocytes were stimulated for 24–96 h with ConA. Nuclear and cytoplasmic protein was extracted and subjected to Western analysis using rabbit antiserum against the C-terminal peptide of Lsh. (E) Time kinetic of cell cycle induction of ConA-activated splenocytes. Splenocytes were stimulated for 2–72 h with ConA and analyzed by flow cytometry for cell cycle.
Flow Cytometric Analysis and Cell Sorting.
Single cell suspensions from thymus, spleen, lymph nodes, and ficolled blood (lympholyte M, Cedarlane Laboratories) were stained and analyzed by flow cytometry using a FACStar Plus (Becton Dickinson). Red blood cells were lysed with ACK buffer (Biofluids, Rockville, MD) and splenocytes were preincubated on ice with 2 μg of normal mouse IgG serum for 5 min to block nonspecific binding by Fc receptors. All antibodies were purchased from PharMingen. Embryonal day 14 thymocytes that were 95% CD44+ were stained with anti-CD25-FITC and sorted into T1 (CD25−) and T2 (CD25+) cell populations. The thymus of 4–8 wk old mice was stained with anti-CD4-PE and anti-CD8-FITC and sorted into double negative, double positive, and single positive cell populations. Population purity was >95%.
Cell Cultures.
For activation of lymphocytes, spleen cells of either C57BL/6J mice or of Lsh−/−Rag2−/− chimeras and littermate controls were cultured at 2.5–3 × 106 cells/ml with either 2 μg/ml ConA (Sigma) or 10 μg/ml lipopolysaccharide (LPS; Sigma) in RPMI medium 1640 (GIBCO/BRL) supplemented with 2 mM glutamine, antibiotics (GIBCO), and 10% fetal calf serum (HyClone). For stimulation with anti-CD3 or anti-CD28, Maxisorb Immuno plates (NalgeNunc) were preincubated with 2 μg/ml of antibody at 4°C overnight. Activated spleen cell supernatant was removed after 24–48 h and tested by ELISA for IL-2 and interferon (IFN; SAIC, Frederick, MD) or antibodies (mAb-based mouse Ig-isotyping kit, PharMingen). To assay proliferation, cells were pulsed for 6 h with 1 μCi of [3H]thymidine (NEN) at the end of the incubation period. For inhibition of Lsh gene induction, splenocytes were cultured with 2 μg/ml of ConA in the presence of 0.2 μM cyclosporine A (Sigma), 1 μg/ml rapamycicn (Sigma), or 10 ng/ml FK506 (kind gift of S. L. Simek, NCI).
Detergent Cell Cycle Analysis.
Cells were washed with 1× PBS, resuspended in 250 μl of detergent buffer (6), and mixed by inversion with an equal volume of staining buffer (50 μg/ml propidium iodide in detergent buffer containing 500 units/ml RNase A; Quiagen, Valencia, CA). After an incubation period of 1 h at room temperature, cells were analyzed by flow cytometry.
Northern and Reverse Transcription–PCR (RT-PCR) analysis.
Northern analysis was performed as described (4). Hybridization was performed overnight using a 32P-random-primed labeled 2.2-kb Lsh cDNA probe (comprising the whole ORF). Five micrograms of total RNA was reverse transcribed using Oligo dT12–18 and Moloney-murine leukemia virus reverse transcriptase (GIBCO). PCR was performed with 1.6 μM of each primer and 0.5 units of AmpliTaq (Perkin–Elmer) according to the manufacturer's recommendation at 94°C for 3 min followed by 30 cycles of 94°C for 1 min, 55–60°C for 1 min, and 72°C for 1 min followed by 7 min extension at 72°C. Annealing temperatures were as follows: cyclin B1 and the ribosomal 18s gene at 60°C; Bax, cyclin D2, and cyclin E at 58°C, and for all others 55°C. PCR reactions were run on 1–2% agarose gels, blotted onto Nytran, hybridized in RapidHyb buffer (Amersham Pharmacia) with a corresponding 32P γ-ATP 5′end-labeled oligonucleotide probe, and visualized by autoradiography. The following murine primer pairs and probes for hybridization were used: cyclinB sense (S) 5′-CAGTGAGTGACGTA GACGCAGA-3′; antisense (AS) 5′-GCACCATGTC GCAGTCCAGCAT-3′; 18s S 5′-TTGATCCTGCC AGTAGCATATGCT-3′; AS 5′-AGTGGGTAATTTGCGCGCC TG-CTG-3′; Bax S 5′-CTACAGGG TTTCATCCAGGATCG-3′; AS 5′-GATGGTCACTGTCTGCCATGTGG-3′; Cyclin D2 S 5′-GTCCG CAGGGCCGTGCCGGACC-3′; AS 5′-GGCAGCTTGCGAAGGATGTGCT-3′; Cyclin E S 5′-GCCCTGGGATGATAATTCAGCA-3′ AS 5′-GGCTTAGACGCCACTT AAGGGC-3′; Bcl-xl S 5′-GTCGAAGAGAATAGGACTGAGG-3′ AS 5′-GTTGAAGCCCT-CCTTGCC TTTC-3′; Bad S 5′-CCTGCACACGCCCTAGGCTTGA-3′ AS 5′-CACCAGGACTGGATAATG CGCG-3′; Bcl-2 S 5′-GGGAAACACCAGAATCAAGT-3′ AS 5′-AGCCAGGAG- AAATCAAACAG-3′; c-myc S 5′-CGCTCCGGGGCGACCTAAGAAGG-3′ AS 5′-GTTCCTGTTGGTGAAG TTCAC-3′; P53 S 5′-GTGTCACGCTTCTCCGAAGAC-3′ AS 5′-CTGGCAGAATAGCTTATTGAG-3′; Fas S 5′-GCATCTCCGAGAGTTTAAAGC-3′ AS 5′-CAAGGGTTCCATGTTCACACG-3′; P21 S 5′-GCCGCCGCGGTGTCAGAGTC-3′ AS 5′-GAAATCTGTCAGGCTGGTCTG-3′; Fas-ligand S 5′-CACTCAAGGTCCATCCCTCTG-3′ AS 5′-ATATCCCTGGTGCCCATGATG-3′; Mdm2 S 5′-TG-TCTGTGTCTACCGAGGGTG-3′ AS 5′-GCCACTAAATTTCTGTAGATC-3′; and Irf-1 S 5′-GAGGA- CCCCAGCATCTCGGGC-3′ AS 5′-GTGTCTCGGCTGGACTTGGAC-3′.
Analysis of V(D)J Recombination.
Genomic DNA was prepared from thymus [for analysis of T cell receptor (TCR) gene rearrangement] and spleen (for analysis of Ig gene rearrangement) from Lsh−/−Rag−/− chimeric animals and Lsh+/+rag−/− controls. Approximately 1 μg of genomic DNA was subjected to PCR analysis, agarose gel electrophoresis, blotting, and specific hybridization with oligonucleotides as described (7). The use of specific oligonucleotides for detection of TCRγ (7), TCRβ (8), TCRα (9), and IgH chain rearrangements (10) have been described previously.
Western Analysis.
Cytoplasmic and nuclear proteins were isolated as described (11). Five to 20 μg of protein extract was separated on an 8% Tris-glycine SDS polyacrylamide gel (NOVEX, San Diego) and blotted onto nitrocellulose. Lsh was detected by using a 1:1,000 dilution of an Ig-purified antiserum (Pierce) of rabbits immunized with a C-terminal peptide of Lsh (Biosynthesis, Lewisville, TX), followed by detection with an anti-rabbit Ig-horseradish peroxidase (HRP) antibody. Detection of HRP was performed using the enhanced chemiluminescence Western blotting detection reagents (Amersham) according to the manufacturer's instructions.
Results
Lsh Expression in Lymphoid Tissue.
Lsh was cloned from a T cell precursor library made from immature thymocytes (>95% CD4−CD8−), a stage undergoing V(D)J recombination (3, 12). As shown by Northern blot in Fig. 1A, Lsh mRNA expression was high in fetal thymocytes from day 14 to 16 of gestation, a population consisting of immature CD4−CD8− pro-T cells. In contrast, Lsh mRNA was low in adult thymus, containing 85–89% CD4+CD8+ cells and only 2–5% CD4−CD8− cells (Fig. 1 A and C). Lsh mRNA was not detectable in unstimulated spleen or lymph node tissue as judged by Northern blot. Thymocytes, in contrast to peripheral lymphocytes in spleen and lymph node, are highly active in V(D)J recombination and cellular differentiation. In addition, fetal thymocytes are rapidly proliferating whereas unactivated spleen and lymph node tissue consist of cells in interphase. Thus, we attempted to correlate Lsh mRNA expression with either cellular differentiation (by cell surface markers), V(D)J recombination, or proliferation.
The earliest T cell precursor stage (pro-T1) is CD25−, and the subsequent stage (pro-T2), prominent between day 14 and 17 of fetal development, expresses CD25. Thus day 14 thymocytes were sorted into CD25+ and CD25− subpopulations and analyzed for Lsh expression by RT-PCR to try to correlate Lsh expression with that of CD25. However, as shown in Fig. 1B, both CD25+ and CD25− populations showed similar amounts of Lsh mRNA. Next we analyzed subsequent stages of thymocyte development in adult thymus for Lsh expression by RT-PCR. However, as shown in Fig. 1B, we found Lsh mRNA expression during all phases of T cell maturation, but lowest in the CD4+ subset. Thus, Lsh mRNA expression was present throughout lymphoid development and could not be strictly correlated to a distinct stage.
Next, we tested the idea of whether Lsh expression correlated with V(D)J recombination of the TCR genes, an event occurring in the fetal thymus and in the CD4−CD8− and CD4+CD8+ subpopulations of the adult thymus. As shown in Fig. 1C, Lsh mRNA was detectable in normally recombining thymocytes but not in either SCID or Rag2−/− recombination-defective thymocytes (13). SCID and Rag2−/− thymocytes are arrested in development at the CD4−CD8− (pro-T3) stage. Stimulation of the pre-TCR by anti-CD3 induced Lsh gene expression in Rag2−/− and SCID thymocytes, as detected by Northern analysis (Fig. 1C). Thus Lsh gene expression can be induced independent of V(D)J recombination. Stimulation of the pre-TCR by anti-CD3 induces a large expansion of Rag2−/− or SCID thymocytes (from 2–3 × 106 to 80–100 × 106) and induces phenotypical differentiation into the CD4+CD8+ T cell stage. However, CD4+CD8+ subsets express rather similar amounts of Lsh mRNA in comparison with CD4−CD8− cells. This suggests that SCID or Rag2−/− thymocytes do not normally express Lsh because they fail to enter the expansion phase after pre-TCR signaling. Thus Lsh expression in thymic development appears to be associated with cell division.
Induction of Lsh in Peripheral Lymphocytes.
Because Lsh expression appeared to correlate with cell division of thymocytes, rather than maturation, we tested whether mature peripheral lymphocytes also could be induced to express Lsh after mitogenic stimulation. As shown in Fig. 2A, Lsh mRNA could be induced in splenocytes by the lectin ConA or by stimulation of the TCR with anti-CD3 antibodies together with the costimulatory signal CD28. Although, the CD28 signal induced a low level of Lsh on its own, CD28 is not obligatory for Lsh expression because CD28−/− mice still show detectable Lsh mRNA levels in ConA activated spleen (data not shown). To test whether ConA induction of Lsh acted via TCR signaling, we used the drugs cyclosporine A and FK506, that block TCR signal transduction. As shown in Fig. 2B, ConA-induced Lsh mRNA expression was blocked by cyclosporine A as well as FK506. This suppression of Lsh was not attributable to a loss of IL-2 production (although FK506 and cyclosporine A both inhibited IL-2 secretion, data not shown) for several reasons: First, addition of IL-2 could not overcome the suppression of FK506 or cyclosporine A (data not shown). Second, IL-2 alone or addition of IL-2 to ConA stimulation did not augment Lsh mRNA expression. Third, rapamycin, which inhibits IL-2 signal transduction, did not influence Lsh gene induction (Fig. 2B). Moreover, IL-7, IFNγ, and IL-4 could not substitute for the TCR and costimulatory signal and TH1 as well as TH2 cells were able to express Lsh mRNA equally well (data not shown). Thus, cytokines did not appear to enhance Lsh mRNA expression, which appears to be dependent on TCR signal transduction and CD28 costimulation.
Because IL-2 promotes cell division and was not required for Lsh induction, this could indicate that Lsh is expressed earlier than the cytokines following T cell activation. However, a time course of Lsh expression, shown in Fig. 2C, indicated that it appeared fairly late, around 48 h after stimulation with the mitogen ConA; IL-2 and other cytokines are produced by 24 h (data not shown). Similarly, Lsh protein was produced late following activation, peaking around 48 h and declining by 96 h (Fig. 2D). As shown, Lsh protein was predominantly localized to the nucleus rather than the cytosol, as would be predicted by its presumed role in chromatin alteration based on its homology with other SNF2/helicase family members. The peak of Lsh mRNA and protein correlated closely with the onset of S phase in ConA activated splenocytes (Fig. 2E). A further correlation of Lsh with cell growth is its constitutive expression in lymphoid cell lines (Fig. 1A and refs. 3 and 4) of both T and B cell lineage. LPS, a B cell mitogen, also induced Lsh mRNA in splenocytes (data not shown). Thus the late appearance of Lsh expression, although inconsistent with a role for Lsh in early T cell activation, suggests a potential role in cell division of T lymphocytes.
Role of Lsh in Lymphoid Development.
To test whether Lsh plays a role in lymphoid development, in either activation or cell cycle, we deleted the Lsh gene in ES cells by homologous recombination (our unpublished results). The targeting vector replaced exon 6 and 7, encoding the ATPase domain I, domain Ia, and part of domain II, with a neomycin casette. The ATPase domain is essential for normal function of SNF2 homologs (14). Premature termination of protein translation by the casette should also block production of the remainder of the protein containing seven helicase domains. As shown in Fig. 3A, Lsh protein of the predicted 87-kDa size could not be detected in activated lymphocytes derived from Lsh−/− embryos.
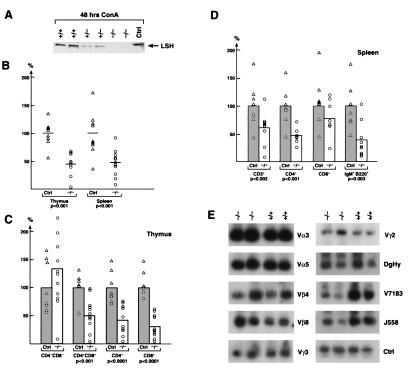
Figure 3.
Effect of Lsh deletion on lymphoid development. (A) Absence of Lsh protein in Lsh−/− splenocytes. Fetal liver cell suspensions of the indicated Lsh genotype were injected into Rag-2−/− recipients and spleens harvested after 4–5 wk. Splenocytes were cultured for 48 h with ConA, nuclear protein extracted, and subjected to Western analysis using rabbit antiserum against the C-terminal peptide of Lsh. (B) Lsh−/− (○, n = 11) chimeric thymus and spleen cell numbers were evaluated and compared to cell numbers generated from + /+ mice (▵, n = 9). No statistically significant difference in thymic or spleen cell numbers was found between + /+ or ± animals. Statistically relevant differences are indicated as P values below. (C) Lsh−/− chimeras (○, n = 11) were analyzed for the thymic subpopulation surface markers CD4 and CD8 by FACs and compared to control +/+ animals (▵, n = 8). Statistically relevant differences are indicated as P values below. (D) Lsh−/− spleen chimeras (○, n = 9) were analyzed by FACs for expression of T cell surface markers CD3, CD4, and CD8 or B cell surface markers B220 and IgM and compared to spleen from + /+ animals (▵, n = 7). Statistically relevant differences are indicated as P values below. (E) Genomic DNA from either thymus or spleen of Lsh−/− or Lsh+/+ chimeric animals was analyzed for V(D)J recombination at the TCRα, β, or γ locus or the IgH chain locus (D-JH4, V7183, J558). The control reactions amplified the Vγ2 gene independent of the recombination event. Only the control reactions for spleen samples are shown.
Deletion of Lsh resulted in a lethal phenotype and the mice die perinatally (our unpublished results). We noted that neonatal Lsh−/− mice had a slightly smaller thymus and the spleen was barely detectable, which could have directly resulted from lymphoid defects, or indirectly via stress effects in these compromised mice. To examine the effect of Lsh on lymphoid development in the absence of other deficiencies (that caused death) we reconstituted Rag2−/− mice, which lack lymphoid cells, with Lsh−/− hematopoietic progenitors. Fetal liver cell suspensions from Lsh−/− embryos (gestational age day 14–18) were used as a source of lymphoid progenitors. At this stage of development, Lsh−/− embryos appeared to be phenotypically normal and at the expected frequency (our unpublished results). In the fetal liver, the frequency of HSA+CD43+ positive cells (from which hematopoietic precursors are derived) was indistinguishable between Lsh−/− or control animals (74.7% B220+ cells in Lsh−/− vs. 78.6% B220+ cells in ± mice and 93.9% HSA+CD43+ cells in Lsh−/− vs. 94% HSA+CD43+ in ± animals). Fetal liver cells were injected into Rag2−/− mice defective in the V(D)J recombinase and lacking endogenous lymphocytes (15). Chimeric animals were analyzed 4–5 wk later for the effect of Lsh deletion on lymphoid development. Thymus and spleen cell numbers were approximately one-half the size in Lsh−/− chimeras compared to controls as shown in Fig. 3B. Throughout the study there was no significant difference in any examined parameter between heterozygous and wild-type mice, although Lsh expression appears to be reduced in heterozygous mice (Fig. 3A).
To test whether the thymic reduction in cell numbers was due to a partial arrest in development or due to a reduction in T cell precursors, we examined the presence of thymic subpopulations in Lsh−/− chimeras. Analysis by flow cytometry showed a relative enrichment in CD4−CD8− T cell precursors from 6.9% in control (Lsh+/− or Lsh+/+) to 23.7% in Lsh−/− animals. The absolute number of each thymic subset was determined, reflecting the reduced number of Lsh−/− thymocytes. As shown in Fig. 3C, only the number of CD4−CD8− T cell precursors were unchanged in the absence of Lsh, whereas the more mature stages (CD4+CD8+ and single positive CD4+ or CD8+ T cells) were reduced by approximately one-half. A detailed analysis of distinct recombination events at the TCR α, β, and γ locus was performed using PCR and a normal rearrangement pattern was observed (Fig. 3E). Cell surface staining for TCR expression (CD3 and TCRαβ chains) confirmed that Lsh−/− thymocytes could generate mature T cells, albeit in smaller numbers. This suggests that Lsh, although not absolutely required for lymphoid development, promotes the differentiation, survival or expansion of thymocytes at the transition from the CD4−CD8− to the CD4+CD8+ stage.
The overall reduction of splenic Lsh−/− lymphocytes was analyzed further for different subsets as shown in Table 1. The proportion of splenic T cells remained about the same as controls (based on staining for CD3 or CD4 T cell markers), although the CD8+ T cell fraction showed a moderate increase. In contrast, the relative proportion of B220+IgM+ B cells were reduced to half that of controls altering the B/T cell ratio in the spleen from 1.4 in control animals to 0.4 in knockout animals. However, mature B cells could be detected as judged by the cell surface expression of IgM and B220. Furthermore, normal rearrangement patterns were observed at the Ig heavy chain locus (Fig. 3E), as had been seen at the TCR locus, indicating that Lsh−/− deficient lymphocytes can perform normal V(D)J recombination.
Table 1.
The effect of Lsh deletion on splenic subpopulations
| Control | −/− | |
|---|---|---|
| 0 | Total spleen, % | Total spleen, % |
| CD3+ | 30.4 ± 12 | 40.5 ± 18 (133%)* |
| B220+ IgM+ | 43.1 ± 11 | 20.6 ± 7.8 (48% P < 0.0001) |
| CD4+ | 15.5 ± 3.5 | 16.6 ± 10.2 (107%) |
| CD8+ | 7.7 ± 2.2 | 14.9 ± 7.3 (193% P < 0.01) |
| Mac1+ | 4.1 ± 0.9 | 6.8 ± 1.8 |
| Mac1+GR1+ | 5.8 ± 1.6 | 20.7 ± 8.3 |
Fetal liver cell suspensions of Lsh−/− and +/+ embryos were injected into Rag-2−/− recipients, and spleens were harvested after 5 wk. Splenocytes were analyzed by FACS for expression of T cell surface markers CD3, CD4, and CD8, or B cell surface markers B220 and IgM. Values represent percentage positive FACS stain of total spleen population and SD. Average numbers summarize four individual experiments with Lsh−/− (n = 11) and control (n = 19). Percentage difference in comparison with control and statistically relevant P values are indicated in parentheses.
Indicates percentage change in comparison to control value that is set as 100%.
The absolute numbers of splenic lymphocyte subpopulations, reflecting the overall decrease in cellularity, are shown in Fig. 3D. T lymphocytes were reduced to 60% in Lsh−/− spleen compared to controls, confirming the block in thymic T cell maturation. In addition to the splenic T cell reduction there was a decrease of peripheral T lymphocytes in the blood of Lsh−/− chimeras (19.3% CD3+ in control vs. 8.8% CD3+ in Lsh−/− animals). In the spleen, the absolute number of B220+IgM+ B cells was reduced to 40% in comparison to controls. A relative reduction in B cell numbers was also detected in lymph nodes (18.9% B220+IgM+ in control vs. 7.2% B220+IgM+ in Lsh−/− animals) and blood (14.7% in control vs. 4.4% in Lsh−/− animals).
Thus deletion of Lsh resulted in a reduction of thymocytes at and beyond the CD4+CD8+ stage, a reduction of peripheral T cells, and a reduction of mature B cells. Therefore, although Lsh was not absolutely required for TCR or Ig gene rearrangement or other lymphoid maturation events, it appears that Lsh controls the expansion or survival of lymphoid cells.
Lsh Is Required for Lymphocyte Proliferation.
Because mature lymphocytes could be generated in the absence of Lsh, we tested lymphocytes for normal function. We first examined the production of cytokines or immunoglobulins from unfractionated spleen cell suspensions. Lsh−/− and control splenocytes were polyclonally activated with ConA and the supernatant tested for IL-2 or IFNγ secretion. As shown in Table 2, Lsh−/− T cells produced both these cytokines in quantities that were not statistically lower than controls, although there was considerable variability between individuals. Thus although the absolute numbers of T cells was reduced, their capacity to produce cytokines was relatively normal, due to the unchanged proportion of T cells in Lsh−/− spleen in comparison with controls (Table 1). This result is also consistent with the previous observation that Lsh is produced in a very late phase of T cell activation, after cytokines are usually produced.
Table 2.
The effect of Lsh deletion on cytokine and Ig secretion
| Protein | Control | −/− |
|---|---|---|
| Cytokines | ||
| IL-2 u/ml | 709 ± 936 (100%*) | 331 ± 211 (51%*) |
| IFNγ ng/ml | 32.8 ± 39 (100%) | 14.1 ± 9.8 (153%*) |
| Immunoglobulins | ||
| IgG1 | 0.41 (100%) | 0.15 (37%) |
| IgG2a | 0.43 (100%) | 0.22 (51%) |
| IgG2b | 0.62 (100%) | 0.32 (52%) |
| IgG3 | 0.48 (100%) | 0.31 (65%) |
| IgM | 0.62 (100%) | 0.49 (79%) |
| IgA | 0.32 (100%) | 0.3 (94%) |
| Igκ | 1.2 (100%) | 0.78 (65%) |
| Igλ | 1.3 (100%) | 0.59 (45%) |
Cytokines. Splenocytes of Lsh−/− chimeric animals were stimulated with ConA for either 24 or 48 h, and the supernatants were tested for IL-2 or IFN-γ activity by ELISA. There was no statistically significant difference between wild-type and heterozygous mice. Data are a summary of four independent experiments with Lsh−/− (n = 10) and Lsh+/− and +/+ animals (n = 28) as control. Immunoglobulins. Splenocytes of Lsh−/− chimeric animals were stimulated with LPS, and supernatant was tested for Ig secretion by ELISA. Control animals are +/+ (n = 2) and +/− (n = 3) compared with Lsh−/− animals (n = 2). Control spleens contained 50.4% B220+IgM+ cells versus 22.8% in Lsh−/− animals.
Percentage variation of knockout supernatants in comparison with control animals as calculated as average variation of four independent experiments.
Analysis of Ig secretion by Lsh−/− splenocytes after stimulation with the polyclonal stimulus LPS showed that levels were comparable to levels of control animals, which is proportional to the reduced B cell numbers in splenic populations. Thus early activation events such as cytokine and Ig secretion were normal in the absence of Lsh.
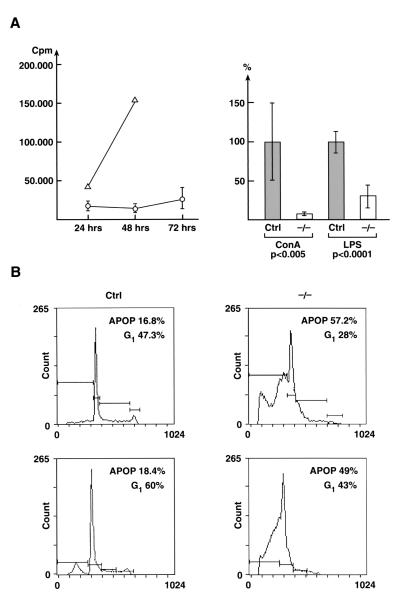
Next we examined the proliferative rate of lymphocytes in response to polyclonal T cell or B cell stimuli. In contrast to cytokine secretion, thymidine incorporation of ConA- activated T lymphocytes was suppressed 10-fold in Lsh−/− splenocytes at 48 h (Fig. 4A). The LPS response, considering the reduced splenic B cell number, showed an almost normal thymidine incorporation (Fig. 4A). The ConA response was then analyzed for cell cycle. As shown in Fig. 4B, ConA-activated cells from Lsh−/− chimeric spleen underwent a much higher rate of apoptosis compared to control spleen. Accordingly, the number of cells in G1 phase was reduced. This result, taken together with the lack of incorporation of thymidine, suggests that activated Lsh−/− T cells undergo programmed cell death before entering S phase of the cell cycle.
Figure 4.
Analysis of proliferation and cell cycle in Lsh−/− chimeras. (A) Effect of Lsh on thymidine incorporation. Fetal liver cell suspensions of the Lsh−/− (○) or control embryos (± ▵) were injected into Rag-2−/− recipients and spleens harvested after 5 wk. Splenocytes were stimulated with ConA for the indicated times (left) or with ConA or LPS for 48 h until thymidine incorporation was measured (right; data summarizes two experiments with Lsh−/− (n = 4) and control (n = 7). (B) Lsh−/− or + /+ spleen cells were subjected to cell cycle analysis after stimulation with ConA for 48 h. Number of cells undergoing apoptosis or in G1 phase of cell cycle are indicated in the upper right corner of two representative Lsh−/− individual mice (n = 10) and two + /+ controls (n = 10).
Thus Lsh may control T cell expression of genes that are required to enter S phase and/or for protection against apoptosis.
Analysis of Gene Expression Involved In Cell Cycle or Apoptosis.
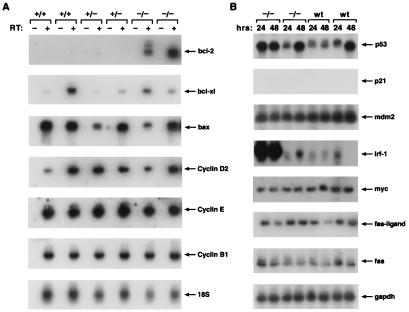
We then examined several candidate genes whose expression might be deregulated in Lsh−/− T cells that could account for cell death before entry into S phase. As shown in Fig. 5A, ConA-activated splenocytes derived from Lsh−/− animals expressed similar amounts of cyclinD1, cyclinB1, and cyclinE mRNA in comparison with control animals. There was no difference in the expression of Fas, Fas-ligand, or c-myc, regulators of cell death, between splenocytes with or without functional Lsh at 24 or 48 h after stimulation (Fig. 5B). Although the expression of p53 and Irf-1, transcription factors that are responsible for DNA-damage induced apoptosis, were variable in distinct animals, there was no significant difference between Lsh wild-type or Lsh-deficient cells. (IRF-1 was highly expressed in two other wild-type splenocyte populations that were analyzed, data not shown). The tumor suppressor gene p53 did not appear to be functionally active in Lsh wild-type or deficient cells, because p21, inducible by active p53, was undetectable (Fig. 5B). Mdm2, binding to and thus counteracting p53 effects, was equally expressed in both populations (Fig. 5B). The expression of the pro-apoptotic gene Bax was similar in Lsh−/−spleen compared to controls (Fig. 5A), whereas mRNA for the pro-apoptotic gene Bad was not detectable in either Lsh−/− or controls (data not shown). Next we analyzed the expression of anti-apoptotic genes Bcl-XL and Bcl-2. Surprisingly Bcl-2 expression, barely detectable in controls, was elevated in the absence of Lsh, whereas Bcl-XL was expressed in similar amounts (Fig. 5A). The high level of Bcl-2 in Lsh−/− T cells did not protect them from apoptotic death, suggesting an effect of Lsh on gene products downstream of Bcl-2 in the death activation cascade.
Figure 5.
Analysis of expression of genes involved in cell cycle or apoptosis. Fetal liver cell suspensions of the indicated Lsh genotype were injected into Rag-2−/− recipients and spleens harvested after 5 wk. Splenocytes were cultured for 48 h with ConA (a) or for 24 and 48 h (b), total RNA extracted, and subjected to RT-PCR analysis for detection of the indicated genes.
Discussion
Lsh is a SNF2/helicase homolog with highest expression in lymphoid tissue in adult animals. Here we report that Lsh−/− hematopoietic precursor cells showed a partial arrest during lymphoid development and that Lsh is required for proliferation of peripheral T lymphocytes.
Because Lsh could be induced by activation through the CD3 complex in SCID and Rag-2−/− thymocytes, this suggests that the pre-TCR signal transduction pathway induces Lsh in immature lymphocytes. The pre-TCR complex includes the TCRβ chain, the pre-Tα and the CD3 complex, and functions to induce CD4CD8 expression as well as intense cell proliferation (16). Consistent with this idea, we observed a reduction in thymic subpopulations at the transition to the double positive stage in Lsh−/− mice leading to reduced numbers of peripheral T cells. However, Lsh is not absolutely required for pre-TCR signaling because mature T cells could be generated in the absence of Lsh.
In contrast, Lsh induction appears to be an essential product of the TCR signal in mature T lymphoytes and crucial for cellular expansion. First, CD3/CD28 costimulation was able to induce expression of the Lsh gene in mature lymphocytes. Second, mature T cells failed to proliferate in the absence of Lsh and undergo apoptosis. Mice with a deletion of IL-7Rα, γc or Jak3 (mediating the IL-7 signal) have also been reported to generate mature lymphocytes that are unable to proliferate in vitro and instead undergo apoptosis (17, 18, 19). Thus Lsh could be connected with signals from cytokines such as IL-7 or could induce cytokines. However, the following points argue against this interpretation: A number of tested cytokines (IL-2, IL-7, IFNγ or IL-4) do not affect Lsh induction and the IL-7Rα−/− thymus shows normal levels of Lsh expression (data not shown). Furthermore, Lsh deletion does not interfere with normal levels of IL-2 or IFNγ secretion. In contrast, Jak3 −/− mice show a severe reduction of secreted IL-2 levels (20). Most importantly, the deletion of γc or Jak3 results in reduced levels of Bcl-2, whereas Bcl-2 levels were elevated in the absence of Lsh (19, 20).
This suggests that Lsh affects lymphocyte proliferation independently of cytokines. It is surprising that Lsh−/− splenocytes undergo apoptosis in the presence of elevated Bcl-2, which blocks or delays apoptotic cell death. However, elevated levels of Bcl-2 have been reported to promote neuronal death mediated by the neurotrophin receptor (21). Alternatively, Lsh could induce a gene that protects from the apoptotic cascade downstream of Bcl-2, for example one of the members of the IAP gene family (inhibitor of apoptosis) (22). An effect of Lsh late in the apoptotic cascade is further supported by the observation that a number of genes involved in the early phase of apoptosis are apparently normally expressed in the absence of Lsh (such as e.g., Fas/Fas-L, p53, Irf-1 or bax). Thus it is possible that Lsh instead affects downstream members of the apoptotic cascade such as the initiator caspase-8 (triggered by death receptors), caspase-9 (triggered by mitochondrial damage), the recently identified caspase-12 pathway (induced by stress in the endoplasmic reticulum), or the “executor” caspases −3, −6, or −7 (23, 24).
To date nothing is known about the biochemical properties of Lsh. However, Lsh protein shows substantial homology to a number of genes involved in chromatin remodeling and transcription rather than to genes involved in DNA repair or recombination, another property of some SNF2/helicase family members (1). Furthermore, there is no evidence that Lsh would participate in V(D)J recombination comprising of site-specific cleavage and a common DNA double-strand break repair mechanism (13). Several genes with high similarity to Lsh extending beyond the helicase domains such as yeast SNF2 (54% similarity), human brahma (BRM) (56%), brahma-related gene-1 (BRG-1) (56%), or Drosophila imitation mating-type switching (ISWI) (53%) have all been shown to disrupt mononucleo-somal arrays in vitro and regulate transcription of specific promoters in vivo (2). However, optimal chromatin remodeling activity in vitro is only achieved in larger protein complexes involving in some cases the presence of up to 11 peptides. Although the chromatin remodeling activity may be primarily connected with “opening” chromatin and transcriptional activation, several reports suggest that SNF2 homologs may also repress chromatin. The CHD family of proteins is thought to inhibit transcription rather than promote (25). Furthermore the SNF2 homolog, Mi-2, is associated with a histone deacetylase complex containing the methyl C-pG-binding protein MBD3. This finding links methylation and histone deacetylation thought to be transciptional silencing to chromatin remodeling (26–28). Consistent with the possibility that loss of Lsh could result in genetic deregulation and overexpression of specific genes is the observation of elevated levels of Bcl-2 mRNA in splenocytes of Lsh −/− mice. Thus, Lsh could possibly participate in processes of chromatin repression rather than “opening”, a hypothesis that deserves to be tested in future.
Acknowledgments
We thank Drs. Scott Durum, Joost Oppenheim, and Dan McVicar for their suggestions on the manuscript and Dr. Scott Durum for helpful discussions. We are grateful to the technical assistance of Rodney Wiles and Louise Finch. This project has been funded in whole or part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. N01-C0–56000.
Abbreviations
- TCR
T cell receptor
- RT-PCR
reverse transcription–PCR
- AS
antisense
- S
sense
- LPS
lipopolysaccharide
- IFN
interferon
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Eisen J A, Sweder K S, Hanawalt P C. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imbalzano A N. Crit Rev Eukaryotic Gene Expression. 1998;8:225–255. doi: 10.1615/critreveukargeneexpr.v8.i3-4.10. [DOI] [PubMed] [Google Scholar]
- 3.Jarvis C D, Geiman T, Vila-Storm M P, Osipovich O, Akella U, Candeias S, Nathan I, Durum S K, Muegge K. Gene. 1996;169:203–207. doi: 10.1016/0378-1119(95)00843-8. [DOI] [PubMed] [Google Scholar]
- 4.Geiman T M, Durum S K, Muegge K. Genomics. 1998;54:477–483. doi: 10.1006/geno.1998.5557. [DOI] [PubMed] [Google Scholar]
- 5.Haks M C, Oosterwegel M A, Blom B, Spits H M, Kruisbeek A M. Semin Immunol. 1999;11:23–37. doi: 10.1006/smim.1998.0153. [DOI] [PubMed] [Google Scholar]
- 6.Telford W G, King L E, Fraker P J. Cell Prolif. 1991;24:447–459. doi: 10.1111/j.1365-2184.1991.tb01173.x. [DOI] [PubMed] [Google Scholar]
- 7.Durum S K, Candeias S, Nakajima H, Leonard W J, Baird A M, Berg L J, Muegge K. J Exp Med. 1998;188:2233–2241. doi: 10.1084/jem.188.12.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlissel M S, Durum S K, Muegge K. J Exp Med. 2000;191:1045–1050. doi: 10.1084/jem.191.6.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roth M E, Holman P O, Kranz D M. J Immunol. 1991;147:1075–1081. [PubMed] [Google Scholar]
- 10.Li Y-S, Hayakawa K, Hardy R R. J Exp Med. 1993;178:951–960. doi: 10.1084/jem.178.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muegge K, West M, Durum S K. Proc Natl Acad Sci USA. 1993;90:4151–4155. doi: 10.1073/pnas.90.9.4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis S M. Ann N Y Acad Sci. 1999;870:58–67. doi: 10.1111/j.1749-6632.1999.tb08865.x. [DOI] [PubMed] [Google Scholar]
- 14.Khavari P A, Peterson C L, Tamkun J W, Crabtree J R. Nature (London) 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 15.Shinkai Y, Rathbun G, Lam K P, Oltz E M, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A M, et al. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 16.Wiest D L, Berger M A, Carleton M. Semin Immunol. 1999;11:251–262. doi: 10.1006/smim.1999.0181. [DOI] [PubMed] [Google Scholar]
- 17.Maraskovsky E, Teepe M, Morrissey P J, Braddy S, Miller R E, Lynch D H, Peschon J J. J Immunol. 1996;157:5315–5323. [PubMed] [Google Scholar]
- 18.Thomis D C, Gurniak E, Tivol A H, Sharpe, Berg L J. Science. 1995;270:794–797. doi: 10.1126/science.270.5237.794. [DOI] [PubMed] [Google Scholar]
- 19.Nakajima H, Shores E W, Noguchi M, Leonard W J. J Exp Med. 1997;185:189–195. doi: 10.1084/jem.185.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baird A M, Thomis D C, Berg L J. J Leukocyte Biol. 1998;63:669–677. doi: 10.1002/jlb.63.6.669. [DOI] [PubMed] [Google Scholar]
- 21.Coulson E J, Reid K, Barrett G L, Bartlett P F. J Biol Chem. 1999;274:16387–16391. doi: 10.1074/jbc.274.23.16387. [DOI] [PubMed] [Google Scholar]
- 22.Deveraux Q L, Reed J C. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner B A, Yuan J. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 24.Mehmet H. Nature (London) 2000;403:29–30. doi: 10.1038/47377. [DOI] [PubMed] [Google Scholar]
- 25.Woodage T, Basrai M A, Baxevanis A D, Hieter P, Collins F S. Proc Natl Acad Sci USA. 1997;94:11472–11477. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wade P A, Gegonne A, Jones P L, Ballestar E, Aubry F, Wolffe A P. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 27.Wade P A, Jones P L, Vermaak D, Wolffe A P. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 28.Jones P L, Wolffe A P. Semin Cancer Biol. 1999;9:339–347. doi: 10.1006/scbi.1999.0134. [DOI] [PubMed] [Google Scholar]