Abstract
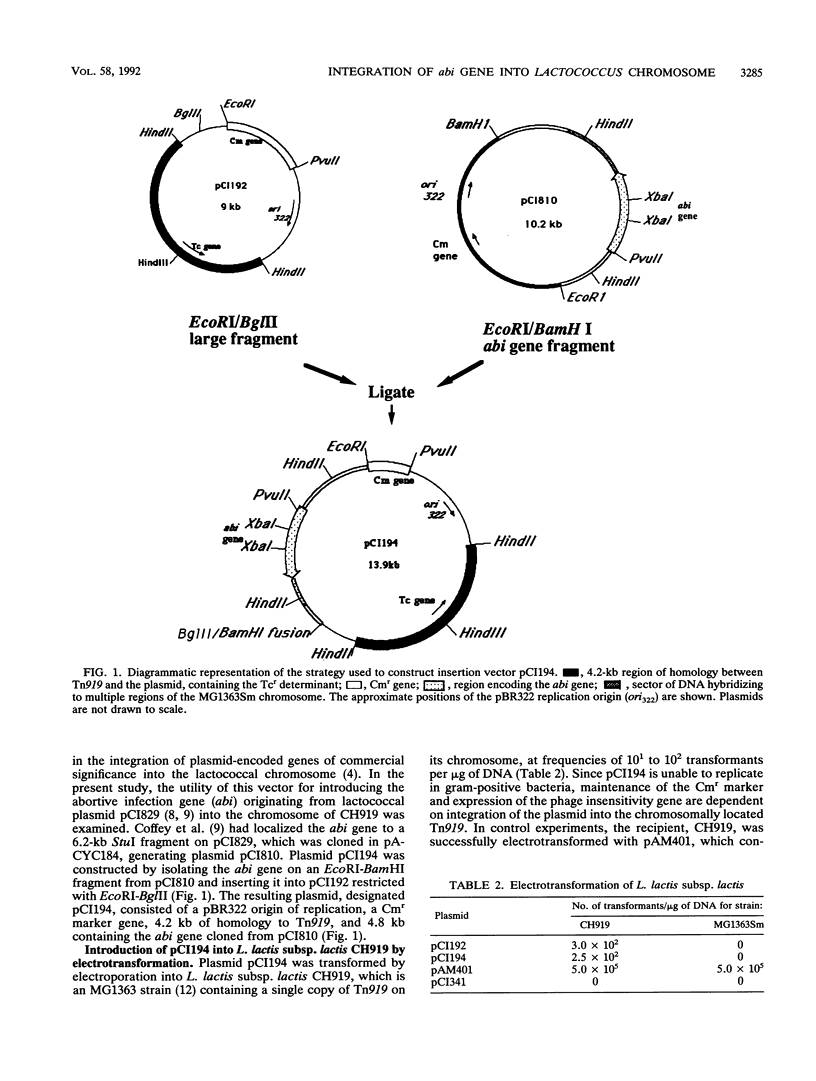
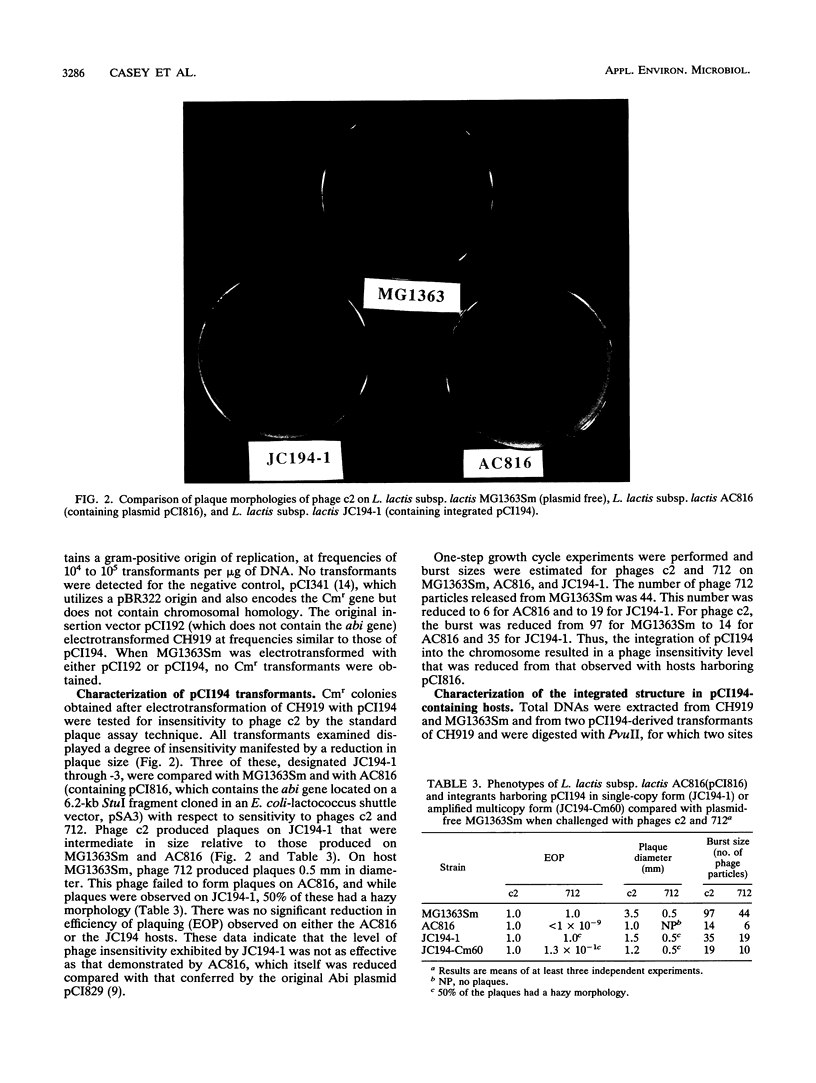
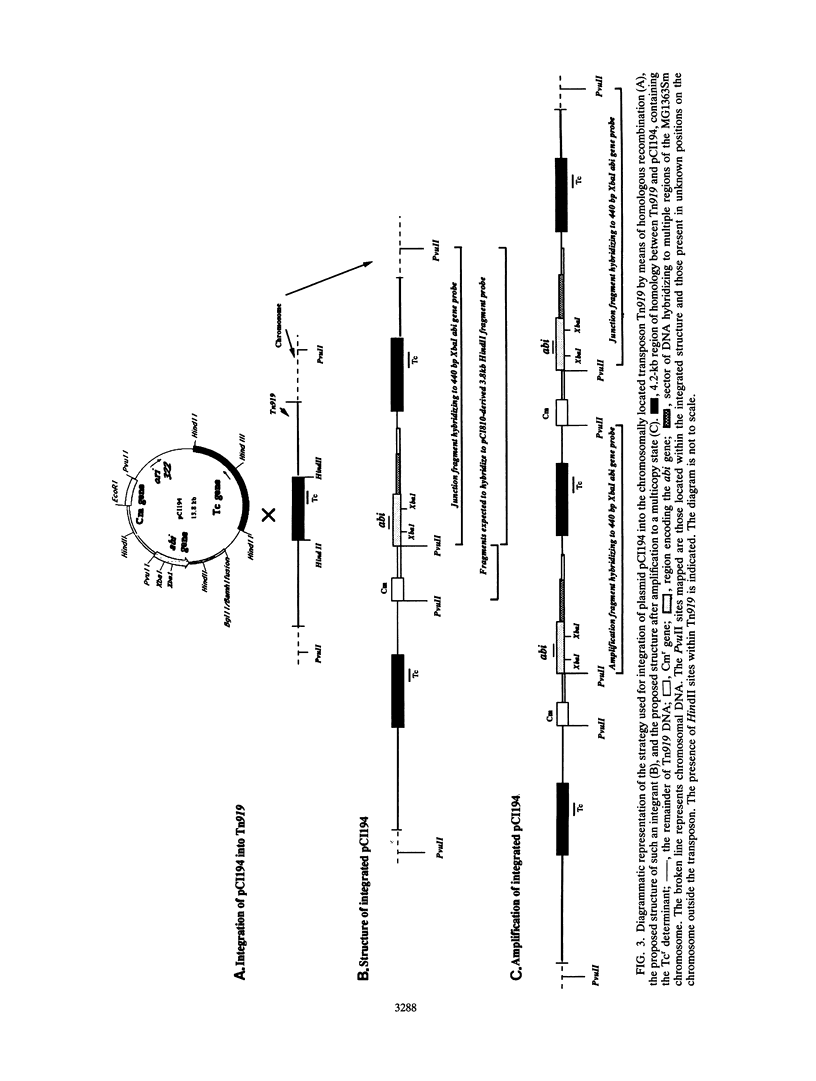
The phage insensitivity gene of lactococcal plasmid pCI829 which encodes an abortive infection defense mechanism (Abi) was inserted into the Lactococcus lactis subsp. lactis CH919 chromosome by utilizing the integration plasmid pCI194, which contains 4.2 kb of homology with the conjugative transposon Tn919. Chloramphenicol-resistant transformants expressed phage insensitivity to the prolate-headed phage c2 and the small isometric-headed phage 712, and hybridization analysis indicated that transformants contained pCI194 integrated in single copy. The level of phage insensitivity expressed by the transformants was reduced from that observed when the abi gene was located on a replicating plasmid, as determined by plaque assay and burst size analysis. Amplification of the integrated structure after growth in increased concentrations of chloramphenicol resulted in an increase in the expression of phage insensitivity. Hybridization analysis revealed that while pCI194 was stably maintained in an integrated state over 100 generations in the absence of selective pressure, the ability to express phage insensitivity was lost. Hybridization analysis also revealed that DNA flanking the abi gene contains homology to the CH919 chromosome.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Casey J., Daly C., Fitzgerald G. F. Chromosomal integration of plasmid DNA by homologous recombination in Enterococcus faecalis and Lactococcus lactis subsp. lactis hosts harboring Tn919. Appl Environ Microbiol. 1991 Sep;57(9):2677–2682. doi: 10.1128/aem.57.9.2677-2682.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin M. C., Chopin A., Rouault A., Galleron N. Insertion and amplification of foreign genes in the Lactococcus lactis subsp. lactis chromosome. Appl Environ Microbiol. 1989 Jul;55(7):1769–1774. doi: 10.1128/aem.55.7.1769-1774.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Yagi Y., Dunny G. M., Schultz S. K. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J Bacteriol. 1974 Jan;117(1):283–289. doi: 10.1128/jb.117.1.283-289.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluzel P. J., Chopin A., Ehrlich S. D., Chopin M. C. Phage abortive infection mechanism from Lactococcus lactis subsp. lactis, expression of which is mediated by an Iso-ISS1 element. Appl Environ Microbiol. 1991 Dec;57(12):3547–3551. doi: 10.1128/aem.57.12.3547-3551.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey A. G., Fitzgerald G. F., Daly C. Cloning and characterization of the determinant for abortive infection of bacteriophage from lactococcal plasmid pCI829. J Gen Microbiol. 1991 Jun;137(6):1355–1362. doi: 10.1099/00221287-137-6-1355. [DOI] [PubMed] [Google Scholar]
- Dodd H. M., Horn N., Gasson M. J. Analysis of the genetic determinant for production of the peptide antibiotic nisin. J Gen Microbiol. 1990 Mar;136(3):555–566. doi: 10.1099/00221287-136-3-555. [DOI] [PubMed] [Google Scholar]
- Fitzgerald G. F., Clewell D. B. A conjugative transposon (Tn919) in Streptococcus sanguis. Infect Immun. 1985 Feb;47(2):415–420. doi: 10.1128/iai.47.2.415-420.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M. J. In vivo genetic systems in lactic acid bacteria. FEMS Microbiol Rev. 1990 Sep;7(1-2):43–60. doi: 10.1111/j.1574-6968.1990.tb04878.x. [DOI] [PubMed] [Google Scholar]
- Gasson M. J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983 Apr;154(1):1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F., Daly C., Fitzgerald G. F. Identification of the Minimal Replicon of Lactococcus lactis subsp. lactis UC317 Plasmid pCI305. Appl Environ Microbiol. 1990 Jan;56(1):202–209. doi: 10.1128/aem.56.1.202-209.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Massey I. J., Klaenhammer T. R. Rapid method to characterize lactococcal bacteriophage genomes. Appl Environ Microbiol. 1991 Jan;57(1):283–288. doi: 10.1128/aem.57.1.283-288.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Miller L. A., Klaenhammer T. R. Nucleotide sequence and distribution of the pTR2030 resistance determinant (hsp) which aborts bacteriophage infection in lactococci. Appl Environ Microbiol. 1990 Jul;56(7):2255–2258. doi: 10.1128/aem.56.7.2255-2258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Romero D. A., McKenney D. S., Finer K. R., Klaenhammer T. R. Localization, cloning, and expression of genetic determinants for bacteriophage resistance (Hsp) from the conjugative plasmid pTR2030. Appl Environ Microbiol. 1989 Jul;55(7):1684–1689. doi: 10.1128/aem.55.7.1684-1689.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannière L., Niaudet B., Pierre E., Ehrlich S. D. Stable gene amplification in the chromosome of Bacillus subtilis. Gene. 1985;40(1):47–55. doi: 10.1016/0378-1119(85)90023-x. [DOI] [PubMed] [Google Scholar]
- Klaenhammer T. R., Sanozky R. B. Conjugal transfer from Streptococcus lactis ME2 of plasmids encoding phage resistance, nisin resistance and lactose-fermenting ability: evidence for a high-frequency conjugative plasmid responsible for abortive infection of virulent bacteriophage. J Gen Microbiol. 1985 Jun;131(6):1531–1541. doi: 10.1099/00221287-131-6-1531. [DOI] [PubMed] [Google Scholar]
- Leenhouts K. J., Gietema J., Kok J., Venema G. Chromosomal stabilization of the proteinase genes in Lactococcus lactis. Appl Environ Microbiol. 1991 Sep;57(9):2568–2575. doi: 10.1128/aem.57.9.2568-2575.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenhouts K. J., Kok J., Venema G. Campbell-like integration of heterologous plasmid DNA into the chromosome of Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1989 Feb;55(2):394–400. doi: 10.1128/aem.55.2.394-400.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- McKay L. L., Bohanon M. J., Polzin K. M., Rule P. L., Baldwin K. A. Localization of Separate Genetic Loci for Reduced Sensitivity towards Small Isometric-Headed Bacteriophage sk1 and Prolate-Headed Bacteriophage c2 on pGBK17 from Lactococcus lactis subsp. lactis KR2. Appl Environ Microbiol. 1989 Oct;55(10):2702–2709. doi: 10.1128/aem.55.10.2702-2709.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polzin K. M., McKay L. L. Identification, DNA sequence, and distribution of IS981, a new, high-copy-number insertion sequence in lactococci. Appl Environ Microbiol. 1991 Mar;57(3):734–743. doi: 10.1128/aem.57.3.734-743.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D. A., Klaenhammer T. R. Characterization of insertion sequence IS946, an Iso-ISS1 element, isolated from the conjugative lactococcal plasmid pTR2030. J Bacteriol. 1990 Aug;172(8):4151–4160. doi: 10.1128/jb.172.8.4151-4160.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero D. A., Klaenhammer T. R. IS946-mediated integration of heterologous DNA into the genome of Lactococcus lactis subsp. lactis. Appl Environ Microbiol. 1992 Feb;58(2):699–702. doi: 10.1128/aem.58.2.699-702.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheirlinck T., Mahillon J., Joos H., Dhaese P., Michiels F. Integration and expression of alpha-amylase and endoglucanase genes in the Lactobacillus plantarum chromosome. Appl Environ Microbiol. 1989 Sep;55(9):2130–2137. doi: 10.1128/aem.55.9.2130-2137.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth R., An F. Y., Clewell D. B. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J Bacteriol. 1986 Mar;165(3):831–836. doi: 10.1128/jb.165.3.831-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]