Abstract
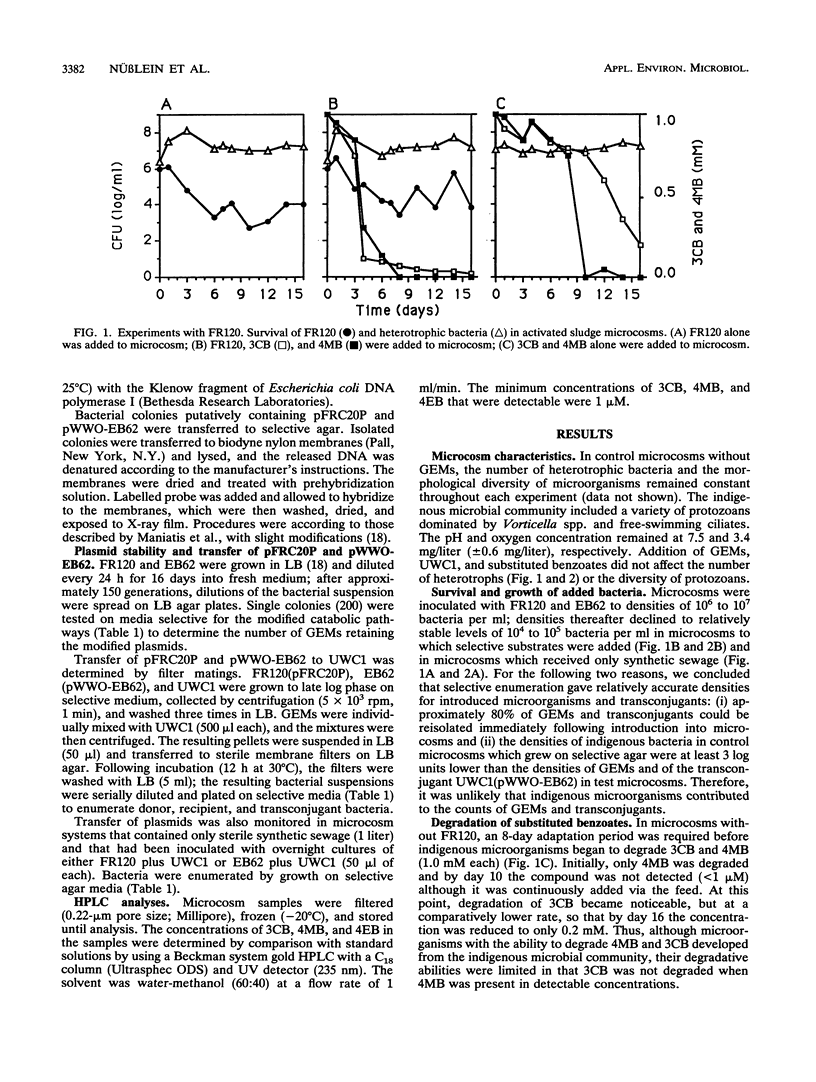
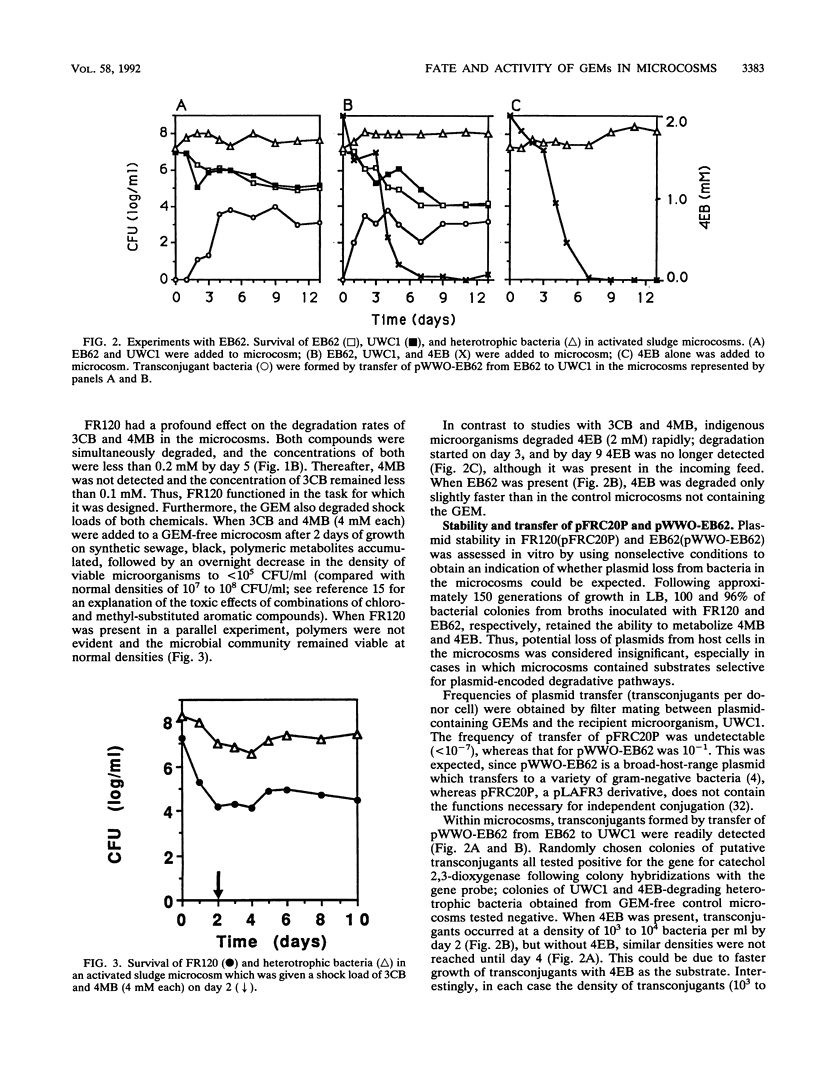
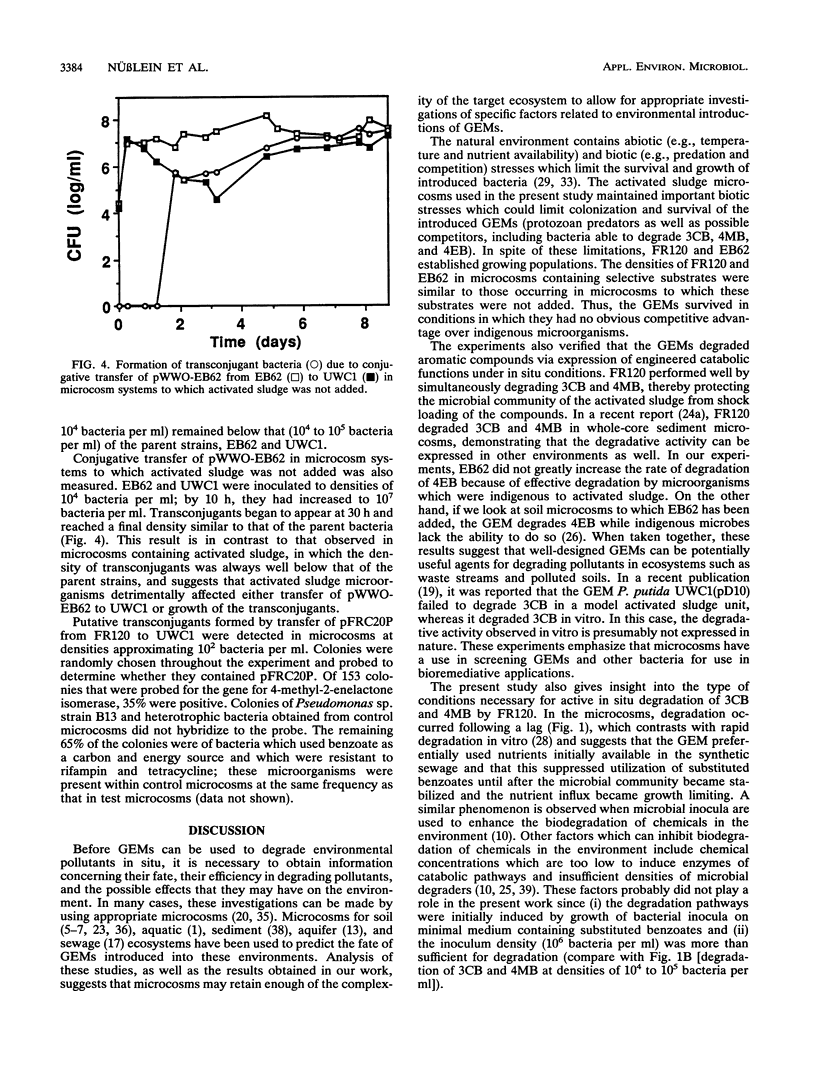
Two genetically engineered microorganisms (GEMs), Pseudomonas sp. strain B13 FR1(pFRC20P) (FR120) and Pseudomonas putida KT2440(pWWO-EB62) (EB62), were introduced into activated sludge microcosms that had the level of aeration, nutrient makeup, and microbial community structure of activated sludge reactors. FR120 contains an experimentally assembled ortho cleavage route for simultaneous degradation of 3-chlorobenzoate (3CB) and 4-methyl benzoate (4MB); EB62 contains a derivative TOL plasmid-encoded degradative pathway for toluene experimentally evolved so that it additionally processes 4-ethyl benzoate (4EB). Experiments assessed survival of the GEMs, their ability to degrade target substrates, and lateral transfer of plasmid-encoded recombinant DNA. GEMs added at initial densities of 10(6) to 10(7) bacteria per ml of activated sludge declined to stable population densities of 10(4) to 10(5) bacteria per ml. FR120 degraded combinations of 3CB and 4MB (1 mM each) following 3 days of adaptation in the microcosms. Indigenous microorganisms required an 8-day adaptation period before degradation of 4MB was observed; 3CB was degraded only after the concentration of 4MB was much reduced. The indigenous microbial community was killed when both compounds were present at concentrations of 4.0 mM. However, in parallel microcosms containing FR120, the microbial community maintained a normal density of viable cells. Indigenous microbes readily degraded 4EB (2 mM), and EB62 did not significantly increase the observed rate of degradation. In filter matings, transfer of pFRC20P, which specifies mobilization but not transfer functions, from FR120 to P. putida UWC1 was not detectable (< 10(-7) transconjugants per donor cell).(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Awong J., Bitton G., Chaudhry G. R. Microcosm for assessing survival of genetically engineered microorganisms in aquatic environments. Appl Environ Microbiol. 1990 Apr;56(4):977–983. doi: 10.1128/aem.56.4.977-983.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Bale M. J., Fry J. C., Day M. J. Transfer and occurrence of large mercury resistance plasmids in river epilithon. Appl Environ Microbiol. 1988 Apr;54(4):972–978. doi: 10.1128/aem.54.4.972-978.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson S., Shapiro J. TOL is a broad-host-range plasmid. J Bacteriol. 1978 Jul;135(1):278–280. doi: 10.1128/jb.135.1.278-280.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentjen S. A., Fredrickson J. K., Van Voris P., Li S. W. Intact soil-core microcosms for evaluating the fate and ecological impact of the release of genetically engineered microorganisms. Appl Environ Microbiol. 1989 Jan;55(1):198–202. doi: 10.1128/aem.55.1.198-202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn E., Hellwig M., Reineke W., Knackmuss H. J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99(1):61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- Goldstein R. M., Mallory L. M., Alexander M. Reasons for possible failure of inoculation to enhance biodegradation. Appl Environ Microbiol. 1985 Oct;50(4):977–983. doi: 10.1128/aem.50.4.977-983.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero M., de Lorenzo V., Timmis K. N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990 Nov;172(11):6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R. K., Sayler G. S., Wilson J. T., Houston L., Pacia D. Maintenance and stability of introduced genotypes in groundwater aquifer material. Appl Environ Microbiol. 1987 May;53(5):996–1002. doi: 10.1128/aem.53.5.996-1002.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knackmuss H. J. Xenobiotic degradation in industrial sewage: haloaromatics as target substrates. Biochem Soc Symp. 1983;48:173–190. [PubMed] [Google Scholar]
- Mancini P., Fertels S., Nave D., Gealt M. A. Mobilization of plasmid pHSV106 from Escherichia coli HB101 in a laboratory-scale waste treatment facility. Appl Environ Microbiol. 1987 Apr;53(4):665–671. doi: 10.1128/aem.53.4.665-671.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure N. C., Weightman A. J., Fry J. C. Survival of Pseudomonas putida UWC1 containing cloned catabolic genes in a model activated-sludge unit. Appl Environ Microbiol. 1989 Oct;55(10):2627–2634. doi: 10.1128/aem.55.10.2627-2634.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvos D. R., Lacy G. H., Cairns J. Genetically Engineered Erwinia carotovora: Survival, Intraspecific Competition, and Effects upon Selected Bacterial Genera. Appl Environ Microbiol. 1990 Jun;56(6):1689–1694. doi: 10.1128/aem.56.6.1689-1694.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton J. M. Genetic engineering and biological detoxification of environmental pollutants. Residue Rev. 1981;78:1–11. doi: 10.1007/978-1-4612-5910-7_1. [DOI] [PubMed] [Google Scholar]
- Pipke R., Wagner-Döbler I., Timmis K. N., Dwyer D. F. Survival and function of a genetically engineered Pseudomonad in aquatic sediment microcosms. Appl Environ Microbiol. 1992 Apr;58(4):1259–1265. doi: 10.1128/aem.58.4.1259-1265.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan M. A., el-Tayeb O. M., Alexander M. Inoculum size as a factor limiting success of inoculation for biodegradation. Appl Environ Microbiol. 1990 May;56(5):1392–1396. doi: 10.1128/aem.56.5.1392-1396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J. L., Duque E., Ramos-Gonzalez M. I. Survival in soils of an herbicide-resistant Pseudomonas putida strain bearing a recombinant TOL plasmid. Appl Environ Microbiol. 1991 Jan;57(1):260–266. doi: 10.1128/aem.57.1.260-266.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J. L., Wasserfallen A., Rose K., Timmis K. N. Redesigning metabolic routes: manipulation of TOL plasmid pathway for catabolism of alkylbenzoates. Science. 1987 Jan 30;235(4788):593–596. doi: 10.1126/science.3468623. [DOI] [PubMed] [Google Scholar]
- Rojo F., Pieper D. H., Engesser K. H., Knackmuss H. J., Timmis K. N. Assemblage of ortho cleavage route for simultaneous degradation of chloro- and methylaromatics. Science. 1987 Dec 4;238(4832):1395–1398. doi: 10.1126/science.3479842. [DOI] [PubMed] [Google Scholar]
- Roszak D. B., Colwell R. R. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987 Sep;51(3):365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saye D. J., Ogunseitan O., Sayler G. S., Miller R. V. Potential for transduction of plasmids in a natural freshwater environment: effect of plasmid donor concentration and a natural microbial community on transduction in Pseudomonas aeruginosa. Appl Environ Microbiol. 1987 May;53(5):987–995. doi: 10.1128/aem.53.5.987-995.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanferlato V. S., Orvos D. R., Cairns J., Lacy G. H. Genetically Engineered Erwinia carotovora in Aquatic Microcosms: Survival and Effects on Functional Groups of Indigenous Bacteria. Appl Environ Microbiol. 1989 Jun;55(6):1477–1482. doi: 10.1128/aem.55.6.1477-1482.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz B., Dahlbeck D., Keen N., Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987 Dec;169(12):5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Top E., Mergeay M., Springael D., Verstraete W. Gene escape model: transfer of heavy metal resistance genes from Escherichia coli to Alcaligenes eutrophus on agar plates and in soil samples. Appl Environ Microbiol. 1990 Aug;56(8):2471–2479. doi: 10.1128/aem.56.8.2471-2479.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevors J. T. Use of microcosms to study genetic interactions between microorganisms. Microbiol Sci. 1988 May;5(5):132–136. [PubMed] [Google Scholar]
- Trevors J. T., van Elsas J. D., van Overbeek L. S., Starodub M. E. Transport of a genetically engineered Pseudomonas fluorescens strain through a soil microcosm. Appl Environ Microbiol. 1990 Feb;56(2):401–408. doi: 10.1128/aem.56.2.401-408.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner-Döbler I., Pipke R., Timmis K. N., Dwyer D. F. Evaluation of aquatic sediment microcosms and their use in assessing possible effects of introduced microorganisms on ecosystem parameters. Appl Environ Microbiol. 1992 Apr;58(4):1249–1258. doi: 10.1128/aem.58.4.1249-1258.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins B. A., Jones S. H., Alexander M. Explanations for the acclimation period preceding the mineralization of organic chemicals in aquatic environments. Appl Environ Microbiol. 1987 Apr;53(4):791–796. doi: 10.1128/aem.53.4.791-796.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]



