Abstract
Chromosomal translocations involving the MLL gene occur in about 80% of infant leukemia. In the search for possible agents inducing infant leukemia, we identified bioflavonoids, natural substances in food as well as in dietary supplements, that cause site-specific DNA cleavage in the MLL breakpoint cluster region (BCR) in vivo. The MLL BCR DNA cleavage was shown in primary progenitor hematopoietic cells from healthy newborns and adults as well as in cell lines; it colocalized with the MLL BCR cleavage site induced by chemotherapeutic agents, such as etoposide (VP16) and doxorubicin (Dox). Both in vivo and additional in vitro experiments demonstrated topoisomerase II (topo II) as the target of bioflavonoids similar to VP16 and Dox. Based on 20 bioflavonoids tested, we identified a common structure essential for topo II-induced DNA cleavage. Reversibility experiments demonstrated a religation of the bioflavonoid as well as the VP16-induced MLL cleavage site. Our observations support a two-stage model of cellular processing of topo II inhibitors: The first and reversible stage of topo II-induced DNA cleavage results in DNA repair, but also rarely in chromosome translocations; whereas the second, nonreversible stage leads to cell death because of an accumulation of DNA damage. These results suggest that maternal ingestion of bioflavonoids may induce MLL breaks and potentially translocations in utero leading to infant and early childhood leukemia.
Approximately 80% of infants (<1 yr of age) with acute myelogenous leukemia (AML) and acute lymphoblastic leukemia (ALL) have chromosome translocations involving the MLL gene (ALL or HRX) at 11q23 (1–3). Moreover, some cancer patients (5–15%) treated with chemotherapeutic agents, such as etoposide (VP16) or doxorubicin (Dox), develop therapy-related leukemia (t-AML, t-ALL) involving MLL (1–3). VP16 and Dox are known inhibitors of eukaryotic topoisomerase II (topo II) (4), an enzyme that alters the DNA topology pivotal for various cell functions by catalyzing double-strand breakage and rejoining of DNA. The presence of topo II inhibitors in cells stabilizes the double-strand breaks and then a covalent topo II–DNA complex can be trapped by using protein denaturants (4, 5). MLL is rearranged with partner genes in 40 different translocations with the most common being the t(4;11), t(6;11), t(9;11), and the t(11;19) (1–3). The translocation breakpoints in MLL localize in an 8.3-kb breakpoint cluster region (BCR); the majority of de novo leukemia breakpoints map to the 5′ half, whereas therapy-related and de novo infant leukemia DNA breakpoints occur predominantly in the 3′ half of the BCR, within a strong scaffold attachment region (6, 7). The finding that the MLL breakpoints of both the therapy-related and infant leukemia patients occur in the same region of the BCR suggests a similar mechanism of DNA damage (7).
It is widely accepted that infant leukemia occurs in utero, based on the presence of leukemia in newborns, in monozygotic twins with leukemia and an identical rearrangement of the MLL gene (8, 9), and by the demonstration of MLL translocations in neonatal blood spots from infants and children who were diagnosed with leukemia (10). Recently, identification of a stillborn with leukemia and an MLL translocation indicates that, in addition to the in utero event, AML can progress very rapidly (11). Putative causative agents for infant leukemia have included maternal exposure to natural or synthetic bioflavonoids during pregnancy (12, 13), some of which inhibit topo II in vitro and which may be similar in their action to chemotherapeutic drugs (5, 14–17). Bioflavonoids are divided into three main groups: flavones, flavanones (2,3-dihydro flavones), and isoflavones, which differ in structure and ring substitutions (18). The inhibitory effect of flavonoids, especially genistein, on human cell growth and tumors has been described (19). In addition, bioflavonoids cross the placenta and are found in fetal tissues (20). In this study, we determined that certain bioflavonoids induce MLL BCR cleavage by inhibiting topo II and thus, by inference maternal ingestion of these bioflavonoids could be associated with the induction of infant leukemia.
Materials and Methods
Cells, Bioflavonoids, and DNA.
BV173 and K562 cell lines (21) were cultured in RPMI medium with 10% FCS (Life Technologies, Gaithersburg, MD). All bioflavonoids, VP16, Dox, ascorbic acid, and bromelain were purchased in high purity from Sigma. One quercetin complex tablet (Solgar, Leonia, NJ) contains 250 mg quercetin, 25 mg citrus complex, and 5 mg rutin. MLL exons 1–34 were isolated from the MLL cDNA, including the 0.74-kb BamHI MLL BCR cDNA containing exons 5–11 (22, 23). To analyze the 5′ terminus of MLL for topo II cleavage sites, the following probes at positions 18,652–19,825, 19,936–20,542, and 32,119–32,442 were PCR-amplified from the cosmid COS20 (ref. 24; Fig. 1).
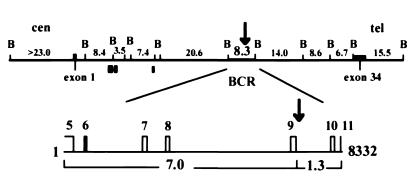
Figure 1.
Diagram of the human genomic MLL. The top map represents the BamHI (B) DNA gene segments with the size (kb) indicated above (22–24). The bottom map represents an enlargement of the 8.3-kb BamHI MLL BCR, which is involved in all MLL chromosome translocations. The entire MLL gene was analyzed for in vivo bioflavonoid- and VP16-induced cleavage sites, centromeric (cen) to telomeric (tel), by use of a series of MLL DNA probes from exons 1 to 34 and genomic fragments corresponding to each BamHI fragment (22–24); these include three centromeric DNA probes (gray boxes) amplified from the cosmid COS20 using PCR at positions 18,652–19,825, 19,936–20,542, and 32,119–32,442 (24). The 7.0- and 1.3-kb cleavage fragments (below the map) representing the BCR were identified with the 0.74-kb cDNA probe (exons 5–11, indicated above map line). The black arrows above both maps represent the colocalizing bioflavonoid- and VP16-induced in vivo topo II cleavage site between nucleotides 6,800 and 7,000 and the DNase I HS site mapped previously (21).
Primary Hematopoietic Cell Cultivation and Differentiation.
The mononuclear fraction was isolated from 50 ml umbilical cord blood, or from peripheral blood collected from healthy adults treated with granulocyte colony-stimulating factor, by density separation on Histopaque (Sigma). A total of 1 × 106 CD34+ cells were isolated by using a CD34+ selection kit (Miltenyi Biotec, Gladbach, Germany) and cultured in McCoy's 5A medium, 12.5% FCS, 12.5% horse serum with human recombinant growth factors (Research Diagnostics, Flanders, NJ), as described previously (25), with some modifications including the addition of stem cell factor (20 ng/ml) and FLT3 ligand (50 ng/ml). After 12 or 14 days, the cells were analyzed with FACS (Becton Dickinson) (25), and the in vivo DNA cleavage assay was performed. For T lymphocyte expansion, the mononuclear cells were isolated by density separation on Histopaque. The remaining red cells were lysed with ACK lysing solution (Sigma), diluted to 0.5 × 106 cells/ml and preincubated overnight in X-VIVO-15 serum-free media (BioWhittaker) with IL-2 (50 ng/ml) and anti-CD3 (OKT-3) antibodies (100 ng/ml) (Biotech, Ratain, NJ). Then, the nonadherent lymphocytes were fractionated from the adherent monocytic cell fraction by decanting into a flask, preincubated with 100 ng/ml anti-CD3 antibodies in PBS. The T lymphocytes were cultured for 12 days and analyzed by FACS, and the in vivo DNA cleavage assay was performed. Genomic DNA isolation and Southern blotting were performed according to Strissel et al. (21).
In Nuclei Topo II DNA Cleavage Reaction.
A 1.6 OD260 of isolated BV173 nuclei were resuspended in a digestion buffer according to Kas and Laemmli (26) for 10 min at 30°C. Then, 2 units or 4 units of purified human topo II (TopoGen, Columbus, OH) were added for 15 min at 30°C. The reaction was started with 1.5 mM ATP, and 50 μM VP16 or 50 μM genistein was added for 5 min at 30°C. The reactions were stopped with 1% SDS and 200 μg/ml proteinase K, and genomic DNA was analyzed.
Topo II-Induced DNA Cleavage Reversibility Reaction.
BV173 cells were incubated with 25 μM and 50 μM VP16 or quercetin for 6 h, and the topo II-induced DNA cleavage was reversed by two different methods. After drug treatment, the cells were either washed and incubated with new media for an additional 2 h or centrifuged and then incubated with 100 mM KCl, 40 mM Tris (pH 7.5), 0.5 mM EDTA (pH 8.0), and 3.0 mM MgCl2 for 15 min at 60°C (27). The reactions were then stopped with 1% SDS and 300 μg/ml proteinase K, and genomic DNA was analyzed.
In Vitro Topo II Relaxation Reaction.
A total of 0.2 μg of supercoiled plasmid DNA pSP72 (Promega), prepared from Escherichia coli, or highly catenated kinetoplast DNA from Crithidia fasciculata (TopoGen) and 1 unit purified human topo II (TopoGen) were incubated at 30°C for 5–20 min in the presence of ATP and bioflavonoids or ATP and chemotherapeutic drugs (14, 15). The reaction products were analyzed on a 1.2% agarose gel and stained with ethidium bromide. The topo II relaxation activity with and without bioflavonoids VP16 and Dox were analyzed by densitometry using the imagequant analysis program (Molecular Dynamics).
Results
Bioflavonoids Cause MLL BCR Cleavage in Primary Progenitor Cells and Cell Lines.
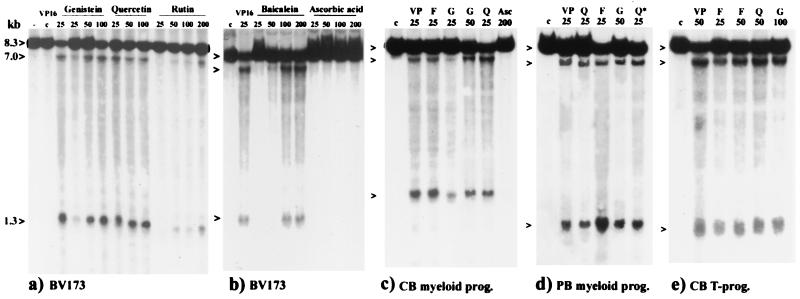
To study the possible role of bioflavonoids in infant leukemia, we examined their effect on DNA cleavage in the MLL gene by using primary hematopoietic progenitor cells and cell lines. CD34+ stem cells and the lymphoid cell fraction were isolated from umbilical cord and from peripheral blood samples of healthy individuals (25). The CD34+ cells were cultured with recombinant growth factors and increased 300-fold with differentiation into both myelocytic and monocytic progenitor cells expressing CD33, CD15, and CD11b. These myelomonocytic cells correspond to the phenotypes of cells observed in infants with AML (28). Both the primary cells as well as two hematopoietic progenitor cell lines (BV173 and K562) were analyzed for DNA cleavage of the MLL gene after culturing for 6 and 16 h with 20 different bioflavonoids: 6 flavones, 5 flavonols, 4 flavanones, 1 flavanol, 4 isoflavones, a commercially available dietary supplement, “quercetin-complex”, as well as the chemotherapeutic agents VP16 and Dox (Table 1), which have been previously shown to induce strong DNA cleavage near exon 9 in the 3′ half of the MLL BCR (refs. 21, 29, and 30; and Fig. 1). Genomic DNA was isolated, digested with BamHI, Southern blotted, and hybridized with a series of probes that scanned the 120-kb of the entire MLL gene (Fig. 1) (22–24). The key results were the following: (i) Only one DNA cleavage site induced by bioflavonoids VP16 and Dox was identified in the entire MLL gene; (ii) The bioflavonoid induced MLL DNA cleavage site mapped to the 3′ half of the 8.3-kb MLL BCR between nucleotides 6,800 and 7,000; it was induced after incubation with 4/6 flavones, 4/5 flavonols, and 2/4 isoflavones, including the dietary supplement quercetin-complex. It also colocalized with the DNA cleavage sites induced by VP16 and Dox (Figs. 1 and 2).
Table 1.
Cleavage of MLL by bioflavonoids and chemotherapeutic drugs
| Name | Origin | Group | in vivo MLL cleavage (μM) | in vitro topo II inhibition |
|---|---|---|---|---|
| Flavone | H | Flavone | 200 | +/− |
| Luteolin | C,R | Flavone | 50 | ++ |
| Apigenin | C,R | Flavone | 100 | ++ |
| Baicalein | H,T | Flavone | 100 | + |
| Rutin* | C,A | Flavone | (200 = 50%) | − |
| Acacetin† | C | Flavone | — | − |
| Fisetin | H | Flavonol | 25 | ++ |
| Kaempferol | C,B | Flavonol | 100 | + |
| Myricetin | B,T | Flavonol | 100 | ++ |
| Quercetin | B,C,A,R | Flavonol | 25 | ++ |
| Quercitrin* | B,C,A,R | Flavonol | — | − |
| Quercetin-co. | D | Flavonol etc. | 25 | ++ |
| Naringenin | C | Flavanone | — | − |
| Naringin* | C | Flavanone | — | − |
| Hesperidin* | C | Flavanone | — | +/− |
| Hesperetin† | C | Flavanone | — | − |
| Taxifolin | C | Flavanol | (200 = 50%) | +/− |
| Genistein | S | Isoflavone | 50 | ++ |
| Genistin* | S | Isoflavone | 100 | + |
| Daidzein | S | Isoflavone | (200 = 50%) | + |
| BiochaninA† | S | Isoflavone | — | − |
| Quer+Fis+Gen | 10 + 10 + 10 | ND | ||
| Quer+VP16 | 10 + 10 | ND | ||
| VP16 | Epipodophyllotoxin | 25 | ++ | |
| Doxorubicin | Anthracycline | 5 | ++ | |
| Ascorbic acid | — | − | ||
| Bromelain | — | − |
The compounds tested are grouped as bioflavonoids, chemotherapeutic (chemo) drugs, or controls. From left to right the first column represents the bioflavonoid common names (*, glycosylated; †, methylated; Que, quercetin; Fis, fisetin; Gen, genistein; Quercetin complex (co), commercially available dietary supplement), and the second column represents some of the main sources of the bioflavonoids (H, herbs; C, citrus; R, root vegetables; T, teas; A, apples; B, berries; D, dietary supplements. The third column states the bioflavonoid group name, and the fourth column represents the bioflavonoid concentration in vivo, which produces the level of MLL BCR cleavage equal to 25 μM VP16 (in bold) after exposure of BV173 cells and primary hematopoietic progenitor cells to the compounds. After hybridization with the MLL BCR cDNA probe, the MLL DNA fragments (8.3 kb germline and the two MLL cleavage products 7.0 kb and 1.3 kb) were analyzed by densitometry. The concentration of each bioflavonoid that produced MLL breakage similar (±5%) to 25 μM VP16 was determined. Note that 200 μM rutin induced only 50% MLL cleavage compared to cells treated with 25 μM VP16. The fifth column represents topo II inhibition in vitro using negative supercoiled plasmid DNA or catenated kinetoplast DNA and purified human topo II: ++, topo II-dependent cleavage activity similar to VP16 or Dox; +, 50% inhibition; +/−, 10–20% inhibition; −, no inhibition. We also defined the BV173 cell line using a FACS and determined this cell line to be a T/B-lymphoid precursor. Our FACS analysis identified 92% viable BV173 cells as: 86% CD71+/38+ and CD71+/38+/10+ but negative for CD45/45RO and CD45RA.
Figure 2.
Southern blots from BV173 (a and b) and human primary progenitor cells (c–e) incubated with bioflavonoids, ascorbic acid, and VP16. Above and from left to right: no addition of drugs (−), DMSO control (C), chemotherapeutic drug VP16 (VP), bioflavonoids as indicated, fisetin (F), genistein (G), quercetin (Q), dietary quercetin-supplement (Q*), and ascorbic acid (Asc). Concentrations (μM) tested are indicated below. Germline (8.3 kb) and the cleavage fragments (7.0 kb and 1.3 kb) are indicated to the left with arrowheads. The FACS results at the time of drug treatment from umbilical cord blood (CB) or peripheral blood (PB) CD34+ ex vivo expansion into myeloid progenitor (prog) cells were (c) 20% CD33+/15+; 51% CD11b+/15+; 14% CD11b+/14+, and 15% propidium iodide+ (PI); (d) 36% CD33+/15+; 33% CD11b+/15+; 26% CD11b+/14+, and 5% PI+; (e) from umbilical cord blood T lymphocyte progenitor cells ex vivo expansion: 27% CD4+/8+; 44% CD4+; 19% CD8+, and 10% PI+.
Quantitative Analyses of Bioflavonoid-Induced MLL DNA Breakage.
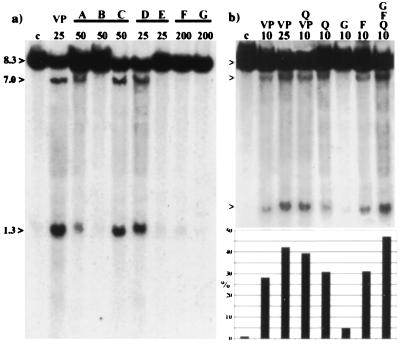
For all cell culture experiments, we used 25 μM VP16 as the standard concentration, which represents the mean plasma concentration measured in cancer patients for the first 6 h after administration (31). The quantity of bioflavonoid- and VP16- induced DNA cleavage products (7.0 and 1.3 kb) in the MLL BCR was compared with the 8.3-kb BamHI germline DNA fragment by use of densitometry of Southern blots hybridized with the MLL BCR cDNA probe (Figs. 1 and 2). The specific concentration of each bioflavonoid that resulted in the same percentage of DNA cleavage in the MLL BCR as 25 μM VP16 is listed in Table 1. The critical observations were the following: (i) the natural flavonols, quercetin and fisetin as well as the commercially available “quercetin-complex”, caused the same level of MLL cleavage as did VP16; (ii) the flavone, luteolin, and the isoflavone, genistein, were 2-fold less active than was VP16; (iii) baicalein (the active ingredient in the Asian herb medicament Shosaiko-to), myricetin, apigenin, and genistin showed 4-fold less activity than did VP16; (iv) flavanones, flavanol, ascorbic acid, and bromelain, the latter two being commonly used in dietary supplements, did not induce MLL cleavage; (v) bioflavonoids with similar ring substitutions, but different ring saturation or geometry induced comparable MLL DNA cleavage (Fig. 3a); (vi) low concentrations of genistein, fisetin, and quercetin in combination, or quercetin and VP16 together demonstrated a cumulative effect for induction of MLL cleavage (Fig. 3b); and (vii) all tested bioflavonoids, whether or not they induced MLL cleavage, caused apoptosis 5–10% over the DMSO control (BV173, 5–10%; primary cells, 10–15%) after 6 and 16 h incubation, using annexin V as a marker (Table 1, and Fig. 2).
Figure 3.
(a) Southern blot of BV173 cells incubated with bioflavonoids with similar ring substitutions, but different ring saturation or geometry (also see Table 1). Above and from left to right: DMSO (C), VP16 (VP), lanes A, B, and C: 4′,5,7-trihydroxyflavonoids (A, apigenin; B, naringenin; C, genistein); D and E: 3,3′4′,5,7-pentahydroxyflavonoids (D, quercetin; E, taxifolin); F and G: 5,7-dihydroxy-4′-methoxyflavonoids (F, acacetin; G, biochaninA). Concentrations (μM) are indicated below. Germline (8.3 kb) and the cleavage fragments (7.0 kb and 1.3 kb) are indicated to the left. (b) Southern blot shows the accumulative activity of bioflavonoids. Above and from left to right: DMSO (C), VP16 (VP), VP16 and quercetin (Q, VP), quercetin (Q), genistein (G), fisetin (F), quercetin, fisetin, and genistein (Q,F,G). Concentrations (μM) are indicated below. Germline and the cleavage fragments are indicated to the left. Below the Southern blot is a densitometry graph, which represents the bioflavonoid cleavage activity in the MLL BCR. Each bar represents the percentage (%) of cleavage and corresponds to the lane above.
We also investigated whether the same structural elements occurred in AF9, a common partner gene of MLL found in 9;11 translocations of infant AML and patients with de novo and t-AML (1). Similar to the MLL-BCR, we colocalized a VP16- and a bioflavonoid-induced DNA cleavage site in intron 7 of AF9, a common site for translocation breakpoints (our unpublished results). Thus, MLL and AF9 may share a common mechanism for gene breakage and rearrangements, which could be induced by bioflavonoids and result in infant leukemia. In contrast, no bioflavonoid-induced in vivo topo II cleavage site was detected in the MLL BCR or in AF9 in VP16-resistant K562 cells, which has been shown by others to have a quantitative reduction as well as a reduced phosphorylation of topo II (32). The resistance of K562 cells to both bioflavonoids and VP16 further supports our proposal that topo II is the target of bioflavonoids.
Topo II Is the Target of Bioflavonoids Causing MLL BCR Breakage.
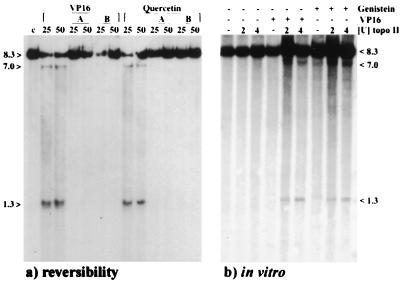
To confirm that cellular topo II was the target of bioflavonoids, we included two in vitro experiments incubating purified human topo II (TopoGen) with DNA or nuclei. First, all bioflavonoids used in this study were assayed for inhibition of DNA decatenation on both supercoiled plasmid DNA and catenated kinetoplast DNA (5, 14–17) by topo II and were then compared with VP16 and Dox. Our study showed that the bioflavonoids inhibited human topo II enzyme activity in vitro similar to VP16 and Dox (Table 1). The bioflavonoids fisetin, myricetin, and quercetin demonstrated inhibition of topo II similar to that of Dox, supporting DNA intercalative activity (5,14–16, and data not shown). Intercalative (e.g., Dox) and nonintercalative anti-tumor drugs (e.g., VP16) as well as the intercalative and nonintercalative bioflavonoids (5, 14–16), all interfere with the breakage-reunion reaction of topo II, causing DNA breakage (4, 27, 33). Second, in vitro experiments were performed with whole nuclei isolated from logarithmically growing BV173 cells (in nuclei topo II assay). After a 5-min incubation of nuclei with purified human topo II and 50 μM of genistein or VP16, cleavage occurred near exon 9 of the MLL BCR, thus confirming our in vivo findings using cultured hematopoietic cells (Fig. 4b).
Figure 4.
Southern blots were hybridized with the 0.74-kb MLL BCR cDNA probe. (a) MLL cleavage induced by 25 μM or 50 μM quercetin and VP16 for 6 h can be reversed using two different methods: A, heat treatment of cells for 15 min at 60°C, immediately after the 6 h drug incubation; or B, an additional incubation of the drug-treated cells in drug-free media for 2 h at 37°C. (b) MLL BCR DNA cleavage induced by genistein and VP16 using an in nuclei topo II assay. Isolated BV173 nuclei were incubated with 2 units or 4 units of purified human topo II and 50 μM VP16 or genistein was added for 5 min at 30°C.
The Bioflavonoid- and VP16-Induced MLL BCR DNA Cleavage Is Reversible.
Two different in vivo drug reversibility experiments were performed to investigate drug–topo II interactions, and to assess whether the MLL BCR cleavage could be reversed. BV173 cells were incubated with 25 and 50 μM VP16 or quercetin for 6 h, then the cells were washed and immediately incubated for 15 min at 60°C in the absence of proteinase K and SDS, or, alternatively, the drugs were removed and the cells were incubated in drug-free medium for an additional 2 h. After DNA isolation and Southern blotting, no MLL breakage was detected with either method (Fig. 4a), indicating the reversibility of VP16- and quercetin-induced DNA breakage. Topo II–DNA cleavable complexes have the unusual property of being unstable at high temperatures (60–65°C), thus causing rapid reversal of the cleavage reaction (4, 27). Other investigators also demonstrated reversibility of genomic DNA cleavage after VP16 and especially genistein treatment (16, 27).
A Common Structure of Topo II-Inhibitory Flavonoids and Isoflavonoids.
The diverse groups of bioflavonoids, the flavones, flavonols, flavanones, flavanols, and isoflavones, are distinguished by different conformational structures and ring substitutions (18). The basic ring system for flavones, flavonols, and isoflavones is planar, whereas the pyran ring of flavanones and flavanols is puckered because of the saturation of the C2–C3 bond. A comparison of the MLL cleavage activity in progenitor cells treated with different groups of bioflavonoids with identical ring substitutions demonstrated the importance of conformational structures (Fig. 3a). MLL breakage with 4′,5,7-trihydroxyflavonoids was detected only with a flavone and isoflavone and not with a flavanone. For 3,3′,4′,5,7-pentahydroxyflavonoids, only a flavonol and not a flavanol induced MLL cleavage. Flavanones and flavanols, as well as glycosylated (except genistin) and methylated bioflavonoids, did not induce MLL breakage (Table 1, and Fig. 3a). Thus, the most potent topo II inhibitors tested in both our in vivo and in vitro experiments were the flavones and flavonols, which are characterized by a planar 7-hydroxy benzopyrone ring with either a 3-or 5-hydroxy substitution and a coplanar 3′-or 4′-hydroxy-2-phenyl ring, and the isoflavones, which are distinguished from flavones by a 4′-hydroxy-3-phenyl ring (Fig. 5).
Figure 5.
The common bioflavonoid minimal structures required for in vivo MLL BCR cleavage and in vitro topo II inhibition. The structure without the shaded side-group represents the flavones (2-phenyl-4H-1-benzopyran-4-one) and with the 3-OH, the flavonols. The benzopyran ring with 3-phenyl substitution (the shaded side-group) represents the isoflavones (3-phenyl-4H-1-benzopyran-4-one). The flavanones (2,3-dihydroflavones) or flavonoids with glycosidic or methoxy groups did not inhibit topo II activity and did not induce MLL BCR DNA breakage.
Discussion
Our research demonstrates that 10 of 20 bioflavonoids induced in vivo DNA cleavage in the MLL BCR between nucleotides 6,800 and 7,000 by topo II inhibition similar to chemotherapeutic agents. The specificity of bioflavonoids for topo II inhibition was directly confirmed by in vitro topo II catalytic (Table 1, and refs. 14–17) and in nuclei topo II DNA cleavage experiments (Fig. 4b). We conclude that some bioflavonoids induce MLL cleavage as actively as VP16 and Dox and represent another class of natural topo II inhibitors. To demonstrate a similarity of the MLL BCR cleavage site in vivo between bioflavonoids and VP16, we have recently analyzed the cleavage site to the nucleotide position using primer extension experiments. Quercetin- and VP16-induced MLL in vivo topo II cleavage sites were staggered by 4 bp (a known feature of topo II DNA cleavage sites; ref. 4) and spanned a region of 47 bp in the high affinity scaffold attachment region of the MLL BCR telomeric of exon 9 (R.S. and P.L.S., unpublished observations). These results confirm our Southern blot analyses of 7.0- and 1.3-kb cleavage products (Figs. 1 and 2). The absence of MLL cleavage in topo II-resistant K562 cells after exposure to bioflavonoids (this report) and VP16 (21, 30, 32) further supports that topo II is the target of bioflavonoids and VP16. Similar evidence was reported by Stanulla et al. (34) with the topo II-resistant CEM-VM15 cell line after a 16-h treatment with VP16.
In reversibility experiments, we demonstrated that bioflavonoid as well as the VP16-induced MLL cleavage can be religated (Fig. 4a). This religation of DNA breakage through the trapped topo II (33) or by DNA repair implies that the cells are not committed to apoptosis. This supports a model of chromosome recombination and cell death by a two-stage pathway of cellular processing of topo II-inhibiting substances proposed by Liu (4). During the first stage, topo II inhibitors, such as VP16, Dox, and bioflavonoids, stabilize the topo II cleavable complexes by forming drug:topo II:DNA ternary complexes on chromosomal DNA. This first stage is reversible by DNA religation or by DNA repair (ref. 4, and Fig. 4a). In contrast, cellular processing of the accumulating ternary complexes activates the second and irreversible stage, which leads to cell death. This hypothesis is supported by studies of cell lines with topo II inhibitors, such as VP16, which resulted in reversible, protein-associated DNA fragments of >300–600 kb in size (35) (first stage of the pathway). Accumulation of these high molecular weight DNA fragments triggered the initiation of apoptotic DNA cleavage, which was irreversible (secondary stage of the pathway) (4, 35).
We propose that only the first, reversible stage of MLL DNA breakage may lead to illegitimate chromosome translocations resulting in leukemia. This notion is supported by studies of VP16-induced, nonhomologous DNA rearrangements using different genes as markers, which showed that cell death did not induce recombinogenesis and that recombinogenesis is not always a lethal event (36). Non-topo II inhibitors such as cytosine arabinoside as well as serum deprivation of cells also resulted in MLL cleavage and cell death (34). In contrast to MLL DNA breakage induced by topo II, and stabilized by topo II inhibitors, the non-topo II inhibitors trigger the initiation of apoptosis and result in apoptotic nucleases cleaving the MLL BCR. The colocalization of topo II and DNase I-induced MLL cleavage indicate that the MLL BCR region between nucleotides 6,800 and 7,000 has an open and accessible conformation susceptible to both topo II and apoptotic nuclease cleavage (21, 34).
In the EUROCLUS study, including infant and childhood leukemia cases from 1980–1989, it was proposed that these leukemias were because of exposure of the fetus or infant to an infectious agent in utero or after birth (37). In contrast to the “infectious-agent” hypothesis, our present study provides the molecular evidence that bioflavonoids induce MLL breakage and strongly supports the conclusion of Ross et al. (13) that dietary bioflavonoids could be a causative agent for infant and possibly childhood leukemia. In a preliminary epidemiological study, it was demonstrated that maternal consumption of topo II inhibitor-containing foods including bioflavonoids led to an approximately 10-fold higher risk of infant AML (38). Based on data from 1971–1980, the incidence of infant ALL and AML for several Asian cities, for example, Hong Kong, Osaka, Miyagi, and Kanagawa, was almost 2-fold higher than in Western countries, where the incidence from 1973–1992 was 37 cases per million per year (United States) (39, 40). This difference in incidence may be because of the high food intake of bioflavonoids in most Asian countries; for example, the Japanese consumption of isoflavonoids in the form of soybeans and soybean products is 1.5–4.1 (genistein) and 6.3–8.3 (genistin) mg per person per day (41). For example, 1.5 μM genistein was detected in human blood plasma after the consumption of 20 g of roasted soybeans. High bioflavonoid concentrations, which persisted for 2–4 days, were also detected in human milk after soy consumption (42, 43). Using primary and BV173 cells, we detected MLL cleavage with low concentration levels of 10 μM quercetin or 10 μM fisetin, which were similar to that with 10 μM VP16 (Fig. 3b). Human plasma levels >1.0 μM of quercetin alone were detected after a quercetin-free diet followed by a single meal, corresponding to 200 mg of quercetin. Moreover, the half-life of quercetin was 25 h, implying that repeated dietary intake of quercetin would lead to much higher blood concentrations (42, 43), similar to those used in this study.
Given the cumulative activity of, for example, quercetin, genistein, and fisetin (Fig. 3b), these compounds in food or dietary supplements like the quercetin complex (one tablet contains 250 mg quercetin) could result in concentrations of bioflavonoids in human blood sufficient to induce DNA cleavage in the MLL BCR. Food or dietary supplement intake of many of the non-topo II inhibitory flavonoids (Table 1) may also be important; for example, the glycosylated flavonol quercitrin is hydrolyzed to the topo II-inhibiting quercetin by intestinal bacteria (44). The conversion of noninhibitory flavonoids could lead to even higher concentrations of topo II-inhibiting flavonoids in the human body. All of the above studies have measured single flavonoid concentrations in different body fluids; however, no data are available concerning the concentrations of multiple flavonoids representing different groups.
However, health benefits of bioflavonoids have been documented. For example, Japan and some other Asian countries show a low mortality rate from prostate cancer when compared with the Western countries. This low mortality rate may be because of high plasma concentrations of isoflavones, e.g., genistein (44-fold), in Japanese men when compared with Finnish men (45), which could act like natural chemotherapeutic agents. The flavonoid quercetin has already been tested in phase I clinical trials in cancer patients. The quercetin concentration in the serum of these patients reached 200–400 μM after administration (46), which is more than 10-fold higher than the level we used for the topo II inhibition studies in vivo and there- fore could lead to MLL breakage resulting in therapy-related leukemia.
In conclusion, although bioflavonoids may be beneficial in certain circumstances, our studies suggest that high dietary intake of bioflavonoids could cause DNA breaks in MLL and possibly in other partner genes by inhibiting topo II. This event could result in chromosome translocations leading to leukemia in adults, children, and particularly infants, analogous to t-AML and t-ALL after cancer chemotherapy.
Acknowledgments
We thank the Cord Blood Bank (Drs. Richard Moldwin and Tom Tyler) and the Leukophoresis Blood Center (Susan Barker and Khadijah James) at the University of Chicago, for providing human umbilical cord and peripheral blood for our studies. We thank Dr. S. Schichman for providing the cosmid COS20 genomic DNA. We thank Drs. N. Zeleznik-Le, U. Storb, F. Olopade, and D. Rowley for their critical comments and discussions. This work was supported in part by the G. Harold and Leila Mathers Charitable Foundation and by the National Cancer Institute Grants CA-42557 and CA-40046 (to J.D.R.).
Abbreviations
- AML
acute myelogenous leukemia
- ALL
acute lymphoblastic leukemia
- VP16
etoposide
- Dox
doxorubicin
- topo II
topoisomerase II
- t-
therapy-related
- BCR
breakpoint cluster region
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.070061297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.070061297
References
- 1.Rowley J D. Annu Rev Genet. 1998;32:495–519. doi: 10.1146/annurev.genet.32.1.495. [DOI] [PubMed] [Google Scholar]
- 2.Ross J A, Davies S M, Potter J D, Robison L L. Epidemiol Rev. 1994;16:243–272. doi: 10.1093/oxfordjournals.epirev.a036153. [DOI] [PubMed] [Google Scholar]
- 3.Canaani E, Nowell P C, Croce C M. Adv Cancer Res. 1995;66:213–234. doi: 10.1016/s0065-230x(08)60255-9. [DOI] [PubMed] [Google Scholar]
- 4.Liu L. Annu Rev Biochem. 1989;58:351–375. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- 5.Markovits J, Linassier C, Fosse P, Couprie J, Pierre J, Jacquemin-Sablon A, Saucier J M, Le Pecq J B, Larsen A K. Cancer Res. 1989;49:5111–5117. [PubMed] [Google Scholar]
- 6.Strissel Broeker P L, Super H G, Thirman M J, Pomykala H, Yonebayashi Y, Tanabe S, Zeleznik-Le N, Rowley J D. Blood. 1996;87:1912–1922. [PubMed] [Google Scholar]
- 7.Cimino G, Rapanotti M C, Biondi A, Elia L, Lo Coco F, Price C, Rossi V, Rivolta A, Canaani E, Croce C M, et al. Cancer Res. 1997;57:2879–2883. [PubMed] [Google Scholar]
- 8.Ford A M, Ridge S A, Cabrera M E, Mahmoud H, Steel C M, Chan L C, Greaves M. Nature (London) 1993;363:358–360. doi: 10.1038/363358a0. [DOI] [PubMed] [Google Scholar]
- 9.Gill Super H J, Rothberg P G, Kobayashi H, Freeman A I, Diaz M O, Rowley J D. Blood. 1994;83:641–644. [PubMed] [Google Scholar]
- 10.Gale K B, Ford A M, Repp R, Borkhardt A, Keller C, Eden O B, Greaves M F. Proc Natl Acad Sci USA. 1998;94:13950–13954. doi: 10.1073/pnas.94.25.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunger S P, McGavran L, Meltesen L, Parker N B, Kassenbrock C K, Bitter M A. Br J Haematol. 1998;103:539–542. doi: 10.1046/j.1365-2141.1998.00994.x. [DOI] [PubMed] [Google Scholar]
- 12.Greaves M F. Lancet. 1997;349:344–349. doi: 10.1016/s0140-6736(96)09412-3. [DOI] [PubMed] [Google Scholar]
- 13.Ross J A, Potter J D, Robison L L. J Natl Cancer Inst. 1994;86:1678–1680. doi: 10.1093/jnci/86.22.1678. [DOI] [PubMed] [Google Scholar]
- 14.Yamashita Y, Kawada S, Nakano H. Biochem Pharmacol. 1990;39:737–744. doi: 10.1016/0006-2952(90)90153-c. [DOI] [PubMed] [Google Scholar]
- 15.Austin C A, Patel S, Ono K, Nakane H, Fisher M. Biochem J. 1992;282:883–889. doi: 10.1042/bj2820883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Constantinou A, Kiguchi K, Huberman E. Cancer Res. 1990;50:2618–2624. [PubMed] [Google Scholar]
- 17.Robinson M J, Corbett A H, Osheroff N. Biochemistry. 1993;32:3638–3643. doi: 10.1021/bi00065a016. [DOI] [PubMed] [Google Scholar]
- 18.Cody V. Prog Clin Biol Res. 1998;280:29–44. [PubMed] [Google Scholar]
- 19.Record I R, Broadbent J L, King R A, Dreosti I E, Head R J, Tonkin A L. Int J Cancer. 1997;72:860–864. doi: 10.1002/(sici)1097-0215(19970904)72:5<860::aid-ijc24>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 20.Schroeder-van der Elst J P, van der Heide D, Rokos H, Morreale de Escobar G, Kohrle J. Am J Physiol. 1998;274:E253–E256. doi: 10.1152/ajpendo.1998.274.2.E253. [DOI] [PubMed] [Google Scholar]
- 21.Strissel P L, Strick R, Rowley J D, Zeleznik-Le N J. Blood. 1998;92:3793–3803. [PubMed] [Google Scholar]
- 22.Nilson I, Lochner K, Siegler G, Greil J, Beck J D, Fey G H, Marschalek R. Br J Haematol. 1996;93:966–972. doi: 10.1046/j.1365-2141.1996.d01-1748.x. [DOI] [PubMed] [Google Scholar]
- 23.Rasio D, Schichman S A, Negrini M, Canaani E, Croce C M. Cancer Res. 1996;56:1766–1769. [PubMed] [Google Scholar]
- 24.Wiedeman L M, MacGregor A, Caldas C. Br J Haematol. 1999;105:256–264. [PubMed] [Google Scholar]
- 25.Smith S L, Bender J G, Maples P B, Unverzagt K, Schilling M, Lum L, Williams S, Van Epps D E. Exp Hematol (Charlottesville, Va) 1993;21:870–877. [PubMed] [Google Scholar]
- 26.Kas E, Laemmli U K. EMBO J. 1992;11:705–716. doi: 10.1002/j.1460-2075.1992.tb05103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsiang Y H, Liu L F. J Biol Chem. 1989;264:9713–9715. [PubMed] [Google Scholar]
- 28.Sorensen P H B, Chen C S, Smith F O, Arthur D C, Domer P H, Bernstein I D, Korsmeyer S J, Hammond G D, Kersey J H. J Clin Invest. 1994;93:429–437. doi: 10.1172/JCI116978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Domer P H, Head D R, Renganathan N, Raimondi S C, Yang E, Atlas M. Leukemia. 1995;9:1305–1312. [PubMed] [Google Scholar]
- 30.Aplan P D, Chervinsky D S, Stanulla M, Burhans W C. Blood. 1996;87:2649–2658. [PubMed] [Google Scholar]
- 31.Relling M V, Yanishevski Y, Nemec J, Evans W E, Boyett J M, Behm F G, Pui C H. Leukemia. 1998;12:346–352. doi: 10.1038/sj.leu.2400928. [DOI] [PubMed] [Google Scholar]
- 32.Ritke M K, Roberts D, Allan W P, Raymond J, Bergoltz V V, Yalowich J C. Br J Cancer. 1994;69:687–697. doi: 10.1038/bjc.1994.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burden D A, Kingma P S, Froelich-Ammon S J, Bjornsti M A, Patchan M W, Thompson R B, Osheroff N. J Biol Chem. 1996;271:29238–29244. doi: 10.1074/jbc.271.46.29238. [DOI] [PubMed] [Google Scholar]
- 34.Stanulla M, Wang J, Chervinsky D S, Thandla S, Aplan P D. Mol Cell Biol. 1997;17:4070–4079. doi: 10.1128/mcb.17.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beere H M, Chresta C M, Alejo-Herberg A, Skladanowski A, Dive C, Larsen A K, Hickman J A. Mol Pharmacol. 1995;47:986–996. [PubMed] [Google Scholar]
- 36.Chen C L, Fuscoe J C, Liu Q, Pui C H, Mahmoud H H, Relling M V. J Natl Cancer Inst. 1996;88:1840–1847. doi: 10.1093/jnci/88.24.1840. [DOI] [PubMed] [Google Scholar]
- 37.Alexander F E, Boyle P, Carli P M, Coebergh J W, Draper G J, Ekbom A, Levi F, McKinney P A, McWhirter W, Magnani C, et al. Br J Cancer. 1998;77:812–817. doi: 10.1038/bjc.1998.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross J A, Potter J D, Reaman G H, Pendergrass T W, Robison L L. Cancer Causes Control. 1996;7:581–590. doi: 10.1007/BF00051700. [DOI] [PubMed] [Google Scholar]
- 39.Parkin D M, Stiller C A, Draper G J, Bieber C A, Terracini B, Young J I, editors. International Incidence of Childhood Cancer, IARC scientific publications, No. 87. Lyon, France: W.H.O.; 1988. [Google Scholar]
- 40.Gurney J G, Ross J A, Wall D A, Bleyer W A, Severson R K, Robison L L. J Pediatr Hematol Oncol. 1997;19:428–432. doi: 10.1097/00043426-199709000-00004. [DOI] [PubMed] [Google Scholar]
- 41.Fukutake M, Takahashi M, Ishida K, Kawamura H, Sugimura T, Wakabayashi K. Food Chem Toxicol. 1996;34:457–461. doi: 10.1016/0278-6915(96)87355-8. [DOI] [PubMed] [Google Scholar]
- 42.Franke A A, Custer L J, Wang W, Shi C Y. Proc Soc Exp Biol Med. 1998;217:263–273. doi: 10.3181/00379727-217-44231. [DOI] [PubMed] [Google Scholar]
- 43.Hollman P C, van Trijp J M, Mengelers M J, de Vries J H, Katan M B. Cancer Lett. 1997;114:139–140. doi: 10.1016/s0304-3835(97)04644-2. [DOI] [PubMed] [Google Scholar]
- 44.Bokkenheuser V D, Shackleton C H L, Winter J. Biochem J. 1988;248:953–956. doi: 10.1042/bj2480953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adlercreutz H, Markkanen H, Watanabe W. Lancet. 1993;342:1209–1210. doi: 10.1016/0140-6736(93)92188-y. [DOI] [PubMed] [Google Scholar]
- 46.Ferry D R, Smith A, Malkhandi J, Fyfe D W, deTakats P G, Anderson D, Baker J, Kerr D J. Clin Cancer Res. 1996;2:659–668. [PubMed] [Google Scholar]