Abstract
Background
Since the creation of “Golden Rice”, biofortification of plant-derived foods is a promising strategy for the alleviation of nutritional deficiencies. Potato is the most important staple food for mankind after the cereals rice, wheat and maize, and is extremely poor in provitamin A carotenoids.
Methodology
We transformed potato with a mini-pathway of bacterial origin, driving the synthesis of beta-carotene (Provitamin A) from geranylgeranyl diphosphate. Three genes, encoding phytoene synthase (CrtB), phytoene desaturase (CrtI) and lycopene beta-cyclase (CrtY) from Erwinia, under tuber-specific or constitutive promoter control, were used. 86 independent transgenic lines, containing six different promoter/gene combinations, were produced and analyzed. Extensive regulatory effects on the expression of endogenous genes for carotenoid biosynthesis are observed in transgenic lines. Constitutive expression of the CrtY and/or CrtI genes interferes with the establishment of transgenosis and with the accumulation of leaf carotenoids. Expression of all three genes, under tuber-specific promoter control, results in tubers with a deep yellow (“golden”) phenotype without any adverse leaf phenotypes. In these tubers, carotenoids increase approx. 20-fold, to 114 mcg/g dry weight and beta-carotene 3600-fold, to 47 mcg/g dry weight.
Conclusions
This is the highest carotenoid and beta-carotene content reported for biofortified potato as well as for any of the four major staple foods (the next best event being “Golden Rice 2”, with 31 mcg/g dry weight beta-carotene). Assuming a beta-carotene to retinol conversion of 6∶1, this is sufficient to provide 50% of the Recommended Daily Allowance of Vitamin A with 250 gms (fresh weight) of “golden” potatoes.
Introduction
Potato (Solanum tuberosum) originated in the highlands of South America, where it has been cultivated for over 2.000 years. Nowadays, potato ranks fourth, among the staple foods of mankind, after wheat, rice and maize. Potato production worldwide stands at 293 million tons, of which 36% in developing countries, and covers more than 18 million hectares (http://www.cipotato.org/market/potatofacts/growprod.htm).
Albeit rich in certain micronutrients, such as vitamin C, cultivated potato is extremely poor in provitamin A. The carotenoid content of tubers in most potato cultivars ranges between 0.5 and 2.5 µg/g FW. The main carotenoids are the xanthophylls lutein and violaxanthin, which are devoid of provitamin A activity. The main provitamin A carotenoid, β-carotene, is present only in trace amounts, from undetectable levels in most cultivars and breeding lines, up to 0.03 µg/g FW [1]. Wild potato species, like Solanum phureja, can reach high carotenoid levels in the tuber, but only a minor fraction of these carotenoids is β-carotene [2] [3]. Due to yield and palatability problems, these species cover only a marginal portion of the worldwide potato production. A botanically distant species, sweet potato (Ipomoea batatas) can reach high carotenoid and beta-carotene levels in the storage root. Sweet potato ranks seventh among staple crops, with 133 million tons produced annually, 117 of which in a single country, China (http://www.cipotato.org/sweetpotato/). Breeding efforts are under way to break the negative genetic linkage between dry matter and β-carotene content in this crop (http://www.harvestplus.org/sweetpotato3.html).
A large effort has taken place, in the past years, for the metabolic engineering of provitamin A in staple foods of plant origin [4]. Perhaps the best known case is that of “Golden Rice”, produced in a joint effort between one of the laboratories authoring the present paper and the laboratory of Ingo Potrykus [5]. Several other cases are known: in the case of canola and of potato, the seed- and tuber-specific overexpression of the bacterial phytoene synthase, CrtB, causes large increases in both total carotenoids and β-carotene in the target tissues [6] [7]. In tomato, constitutive overexpression of the bacterial phytoene desaturase/isomerase, CrtI, causes β-carotene accumulation and a slight decrease in total carotenoids [8]. In the case of “Golden Rice”, a mini-pathway driving synthesis of β-carotene from geranylgeranyl diphosphate (Figure 1A) has been introduced in rice endosperm (reviewed in [9]). The first step, phytoene synthase (PSY), was of plant origin and was expressed under the control of the endosperm-specific glutelin promoter [5]. The desaturation steps (Figure 1A) were performed by a multifuctional enzyme of bacterial origin, CrtI, fused to the RbcS transit peptide and put under the control of the constitutive 35S promoter [5] [10]. Lycopene cyclase was shown to be dispensible due to sufficient expression of the corresponding endogenous enzyme [5] [11]. Later versions of Golden Rice employed endosperm-specific promoters for all carotenoid transgenes, PSY genes from cereals, and have highly increased β-carotene levels (up to 31 µg/g DW) [12]. Codon-optimized versions of the CrtI gene have been also tested [13].
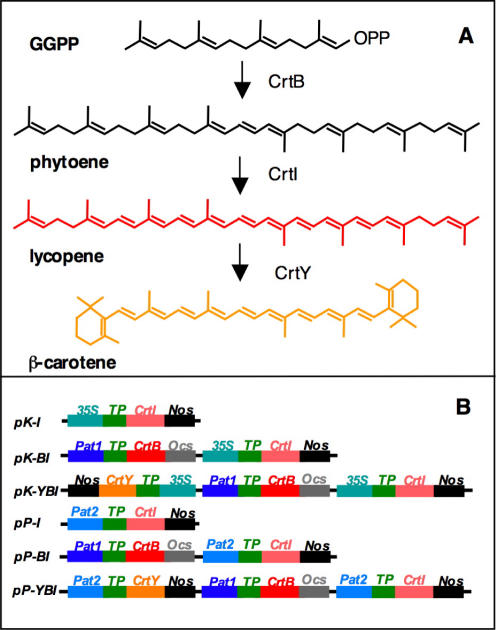
Figure 1. Strategy for the enhancement of the carotenoid content of potato tubers.
A: Biosynthetic pathway catalyzed by the CrtB-I-Y genes. B: Schematic representation of the constructs utilized for the transformation experiments. TP: RbcS transit peptide. Nos and Ocs: Nopaline synthase and Octopine synthase polyadenylation sequences; 35S: Constitutive CaMV 35S promoter; Pat1 and Pat2: Tuber-specific patatin promoters. For details, see Materials and Methods.
In this paper we report the results of a systematic investigation, aimed at verifying which is the optimal combination of promoters and transgenes, able to maximally increase the provitamin A content of potato tubers without affecting vegetative characteristics.
Results and Discussion
The Erwinia genes encoding phytoene synthase (CrtB), phytoene desaturase/carotene isomerase (CrtI) and lycopene beta-cyclase (CrtY) mediate the conversion of geranylgeranyl diphosphate (GGPP) into β-carotene (Figure 1A). The ORFs were fused to the RbcS transit peptide to direct the encoded protein into plastids (Figure 1B). A synthetic CrtI gene was used, whose codon usage has been optimized to match that of plants [13]. In all the constructs, the first gene in the pathway (CrtB) was put under the control of a tuber-specific Pat1 promoter, to avoid dwarfism caused by constitutive expression of phytoene synthase [14]. This strategy has been already shown to result in consistent increases of tuber carotenoid and β-carotene content [7]. Genes encoding later steps in the pathway (CrtI and/or CrtY) were fused to the 35S promoter in the pK series, or to the tuber-specific Pat2 promoter in the pP series of constructs. Six constructs, containing different transgene-promoter combinations, were tested (Figure 1B).
Plasmids belonging to the pK series caused lower transformation efficiencies than those belonging to the pP series, indicating that the constitutive expression of CrtY and/or CrtI interfered, to some extent, with the successful establishment of transgenosis. Within the same series, different constructs gave again different efficiencies, in the following order: CrtI> CrtYBI>CrtBI (Table 1).
Table 1. Transformation frequencies.
| Construct | % regeneration | % PCR-positive regenerants | % Transgenosis |
| pK-I | 61 | 66 | 40 |
| pK-BI | 24 | 41 | 10 |
| pK-YBI | 47 | 37 | 17 |
| pP-I | 73 | 71 | 52 |
| pP-BI | 29 | 38 | 11 |
| pP-YBI | 64 | 57 | 36 |
The % of leaf discs giving at least 1 regenerant after 8 weeks on kanamycin is shown in the second column. The % of PCR-positive shoots containing the transgene are shown in the third column. The % transgenosis (fourth column) indicates the % of leaf disks giving at least 1 PCR-positive regenerant.
The carotenoid content of tubers and leaves of minimum of 9 independent transgenic lines for each construct was analyzed spectrophotometrically, for a total of 86 lines (Figure 2). As can be seen, in plants harboring the pK constructs there is an inverse correlation between tuber and leaf carotenoid content: plants with higher tuber carotenoids have lower leaf carotenoids, and vice versa (Figure 2A). Plants harboring the pP constructs, in which the CrtY and/or CrtI genes are under the control of the Pat2 promoter, do not show significant changes in leaf carotenoid content (Figure 2B). For most of the constructs, the maximum increase in tuber carotenoids is 3-fold. This is less than what has been obtained with transformation with Pat:CrtB alone (4–7 fold) [7].
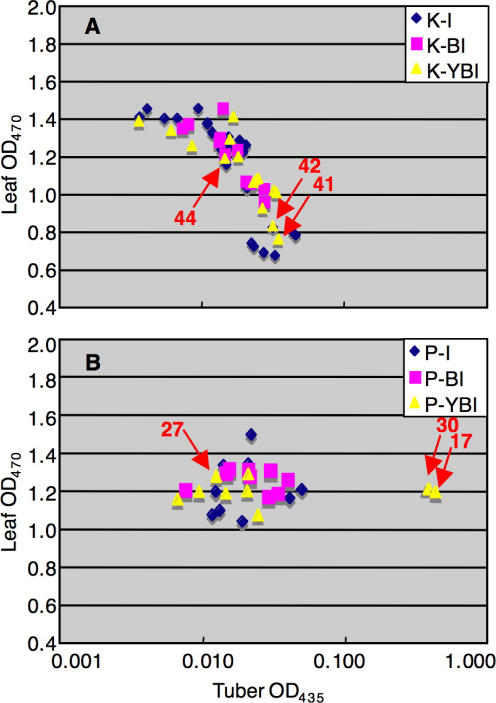
Figure 2. Spectrophotometric quantitation of tuber and leaf carotenoids in transgenic lines.
A: Lines transformed with the pK constructs (see Figure 1B). B: Lines transformed with the pP constructs (see Figure 1B). Data are the average of 4 independent tubers from 2 independent plants. Lines submitted to HPLC and Real Time RT-PCR analysis (Tables 2–3 and Figure 4) are indicated by arrows.
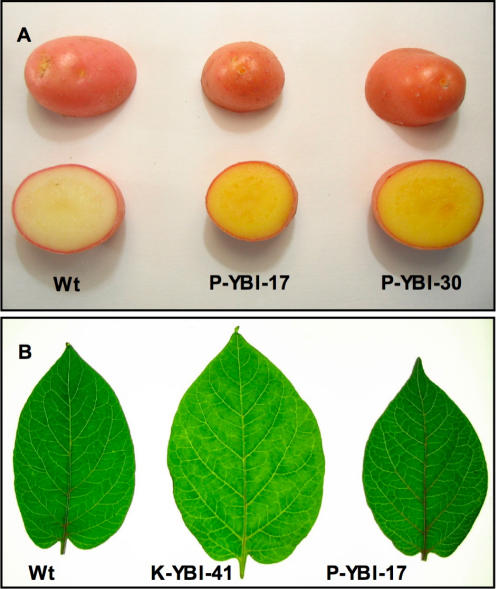
The pP-YBI construct, in which the whole mini-pathway is expressed under tuber-specific promoter control, stands out from all the other constructs: in two lines (P-YBI 17 and 30), tuber carotenoid content is increased approx. 20-fold, without appreciable effects on leaf carotenoid content (Figure 2B). This increase in tuber carotenoids is much higher than what has been obtained previously with Pat:CrtB alone [7]. Tubers of these two lines have a “golden” color (Figure 3A), while their leaves have a normal morphology and pigmentation (Figure 3B). To the opposite, leaves of the pK lines with reduced carotenoid content showed signs of chlorosis (Figure 3B).
Figure 3. Tuber and leaf phenotypes of transgenic lines.
A.Tuber phenotypes. B.Leaf phenotypes, viewed in transmitted light. The difference in size of the middle leaf is not representative.
Our interpretation of these data is that, in the pK constructs, in which the CrtY and/or CrtI genes are under the control of the 35S promoter, leaf expression of one of the introduced transgenes interferes with leaf carotenogenesis. As a consequence, high expressing lines are counterselected and tuber carotenoid levels show only modest (up to 3-fold) increases. This hypothesis was verified via Real Time RT-PCR on selected transgenic lines (indicated by arrows in Figure 2). The results (Table 2) indicate that the two lines (pK-YBI 41 and 42) showing the highest reduction in leaf carotenoids also show the highest expression of the CrtI transcript in leaves, while all the other lines show relatively low expression levels in the same tissue. Changes in carotenoid composition have been observed in leaves expressing the wild-type CrtI gene under 35S promoter control [15] [8]. In the present case, use of a codon-optimized CrtI gene probably leads to much higher levels of CrtI protein [13], and thus the potential for interference is higher. In agreement with this model, the pK-YBI 44 line, showing very low levels of transgene expression, has wild-type leaf and tuber carotenoid levels.
Table 2. Transgene expression in leaves and tubers.
| CrtB | CrtI | CrtY | ||||
| Leaf | Tuber | Leaf | Tuber | Leaf | Tuber | |
| Wild-type | nd | nd | nd | nd | nd | nd |
| pK-YBI 41 | 0.0006±0.0001 | 0.0026±0.0004 | 0.2094±0.0101 | 0.1349±0.0195 | 0.0777±0.0148 | 0.024±0.0023 |
| pK-YBI 42 | 0.0003±0.0002 | 0.0025±0.0004 | 0.1914±0.0065 | 0.0865±0.0166 | 0.0345±0.0109 | 0.0118±0.0028 |
| pK-YBI 44 (NE) | nd | nd | 0.0006±0.0002 | 0.0013±0.0001 | 0.0003±0.0001 | 0.0002±0.0001 |
| pP-YBI 17 | 0.0004±0.0002 | 1.1906±0.0882 | 0.0015±0.0004 | 12.6922±2.6657 | 0.0006±0.0001 | 4.6391±0.0793 |
| pP-YBI 30 | 0.0005±0.0002 | 1.1142±0.2328 | 0.0029±0.0007 | 3.5258±0.2289 | 0.0006±0.0002 | 2.8757±0.1778 |
| pP-YBI 27 (NE) | nd | nd | 0.0003±0.0001 | 0.0018±0.0001 | nd | 0.0006±0.0003 |
Values are normalized with respect to the β-tubulin transcript. For each construct, two lines with significant carotenoid changes and one “non expressor” line (NE) are shown.
The carotenoid composition of tubers and leaves from selected lines was analyzed via diode array HPLC. In different harvests, only minor differences in carotenoid content were observed, maintaining the fold increase in carotenoid content over the wild-type and the relative distribution of individual carotenoids. In table 3, we show the mean and standard deviation of carotenoid content from two different harvests.
Table 3. HPLC analysis of tuber and leaf pigments (µg/g dry weight).
| TUBERS | |||||||||
| Line | Phyt | Alpha | Lutein | Beta | Zea+Anthera | Viola | Neo | Other | Total |
| Wild-type | 0.0 | 0.0 | 1.0±0.3 | 0.013±0.002 | 1.9±0.5 | 0.7±0.2 | 0.6±0.2 | 1.5±0.6 | 5.8±1.8 |
| pK-YBI 41 | 0.0 | 0.0 | 2.2±0.6 | 1.8±0.3 | 6.6±2.0 | 0.6±0.2 | 1.4±0.4 | 4.2±0.8 | 16.5±5.2 |
| Fold Variation | – | – | 2.2 | 135.3 | 3.5 | 0.8 | 2.3 | 2.8 | 2.8 |
| pK-YBI 42 | 0.0 | 0.0 | 2.0±0.8 | 1.8±0.3 | 5.8±2.5 | 0.6±0.3 | 2.3±0.8 | 2.6±1.4 | 15.2±6.8 |
| Fold Variation | – | – | 2.0 | 142.6 | 3.1 | 0.9 | 3.8 | 1.7 | 2.6 |
| pK-YBI 44 | 0.0 | 0.0 | 0.6±0.1 | 0.0 | 1.8±0.4 | 0.4±0.1 | 0.3±0.1 | 1.8±0.4 | 4.9±0.9 |
| Fold Variation | – | – | 0.6 | – | 0.9 | 0.5 | 0.5 | 1.2 | 0.8 |
| pP-YBI 17 | 13.8±6.1 | 6.2±2.4 | 23.1±5.2 | 47.4±18.0 | 11.0±3.7 | 5.6±1.8 | 5.7±2.3 | 1.7±0.6 | 114.4±41.5 |
| Fold Variation | – | – | 23.1 | 3643.3 | 5.8 | 7.9 | 9.5 | 1.1 | 19.7 |
| pP-YBI 30 | 19.5±1.4 | 6.1±2.6 | 30.0±7.3 | 26.4±4.7 | 6.2±1.4 | 22.3±5.7 | 0.5±0.2 | 2.1±1.4 | 112.9±24.9 |
| Fold Variation | – | – | 29.9 | 2031.9 | 3.3 | 31.8 | 0.8 | 1.4 | 19.5 |
| pP-YBI 27 | 0.0 | 0.0 | 0.9±0.1 | 0.0 | 3.0±0.4 | 0.4±0.1 | 0.6±0.2 | 1.5±0.1 | 6.4±0.9 |
| Fold Variation | – | – | 0.9 | – | 1.5 | 0.5 | 0.9 | 1.0 | 1.1 |
| LEAVES | |||||||||
| Line | Chl a | Chl b | Total Chl | Lutein | Beta | Viola | Neo | Other | Total |
| Wild-type | 13846±2038 | 4343±642 | 18190±2680 | 1833±1 | 1165±1 | 414±1 | 617±1 | 308±1 | 4337±1 |
| pK-YBI 41 | 8269±2095 | 2131±99 | 10400±2194 | 1177±418 | 639±166 | 238±51 | 446±106 | 178±16 | 2679±757 |
| Fold Variation | 0.6 | 0.5 | 0.6 | 0.6 | 0.5 | 0.6 | 0.7 | 0.6 | 0.6 |
| pK-YBI 42 | 8140±1123 | 2768±221 | 962±98 | 962±98 | 524±66 | 198±23 | 368±37 | 193±89 | 2245±313 |
| Fold Variation | 0.6 | 0.6 | 0.6 | 0.5 | 0.4 | 0.5 | 0.6 | 0.6 | 0.5 |
| pK-YBI 44 | 14169±3241 | 4619±391 | 18788±3821 | 2037±1 | 1292±1 | 398±1 | 631±1 | 337±1 | 4695±1 |
| Fold Variation | 1.0 | 1.1 | 1.0 | 1.1 | 1.1 | 1.0 | 1.0 | 1.1 | 1.1 |
| pP-YBI 17 | 15208±1593 | 4800±581 | 20009±2174 | 2515±393 | 1476±230 | 459±93 | 691±125 | 358±76 | 5500±919 |
| Fold Variation | 1.1 | 1.1 | 1.1 | 1.4 | 1.3 | 1.1 | 1.1 | 1.2 | 1.3 |
| pP-YBI 30 | 12674±3377 | 3865±716 | 16539±4094 | 1883±374 | 869±169 | 310±45 | 537±121 | 245±35 | 3845±745 |
| Fold Variation | 0.9 | 0.8 | 0.9 | 1.0 | 0.7 | 0.7 | 0.9 | 0.8 | 0.9 |
| pP-YBI 27 | 13297±3016 | 4102±152 | 17399±4120 | 1612±106 | 981±137 | 348±32 | 654±73 | 287±73 | 3883±422 |
| Fold Variation | 1.0 | 0.9 | 1.0 | 0.9 | 0.8 | 0.8 | 1.1 | 0.9 | 0.9 |
Carotenoid composition was measured via diode array HPLC (see Methods) on a minimum of 8 different tubers or leaves from 4 different plants, belonging to 2 different harvests. Fold variation with respect to the wild-type is reported for each carotenoid compound and for each line.
As can be seen, in lines pK-YBI 41 and 42 tuber carotenoids increase, respectively, 2.8 and 2.6-fold. The single carotenoid showing the highest increase is the final product of the introduced mini-pathway, β-carotene, that increases >130-fold. Leaf carotenoid content shows instead a significant decrease, affecting in a comparable fashion all carotenoid species present, as well as Chl a and Chl b.
“Golden” tubers from lines pP-YBI 17 and 30, in which all three transgenes are under Patatin promoter control, show extremely high carotenoid levels, >110 µg/g dry weight. This is at least 3-fold higher than the highest carotenoid content previously reported in potato tubers, including Pat:CrtB engineered plants [7], wild accessions like S. phureja [2], cultivars carrying the Orange flesh gene [1] or transgenic plants carrying the cauliflower Or gene [16]. This vast increase in tuber carotenoid content is associated with very high levels of expression (1 to 12-fold tubulin) of the CrtB-I-Y transgenes in the tuber, but very low levels of expression of the same transgenes in the leaves (Table 2). Leaf carotenoid content is not affected in “golden” lines (Table 3). β-carotene is the carotenoid species showing the largest increase in “golden” tubers: it increases >3600-fold, to 47 µg/g dry weight, in line pP-YBI 17. This is approx. 5-fold higher than what reported previously [7] and is also the highest β-carotene content reported for the 4 major staple foods of plant origin (wheat, maize, rice, potato), including “Golden Rice 2”, which contains up to 36 µg/g dry weight total carotenoids and 31 µg/g dry weight β-carotene [12]. β-carotene is not the only carotenoid species showing massive increases in these two lines: phytoene, which is undetectable in the Wt line, increases up to 19 µg/g dry weight, indicating that, in spite of the high levels of expression of the codon-optimized CrtI transgene, phytoene desaturation is still a rate-limiting step. This finding differs from what has been reported in canola seeds, where overexpression of CrtI in conjunction with CrtB decreases phytoene to trace levels, with respect to overexpression of CrtB alone; another difference between canola and potato is that overexpression of CrtY in conjunction with the other two genes does not bring a further increase in total carotenoids [6] [17].
Other carotenoid species show also massive increases in “golden” tubers: α-carotene, which is undetectable in the Wt line, reaches 6 µg/g DW, bringing total provitamin A carotenoids (α- and β-carotene) to 53 µg/g dry weight in line pP-YBI 17. The xanthophylls lutein and violaxanthin, derived, respectively, from α- and β-carotene, increase up to 30-fold (Table 3).
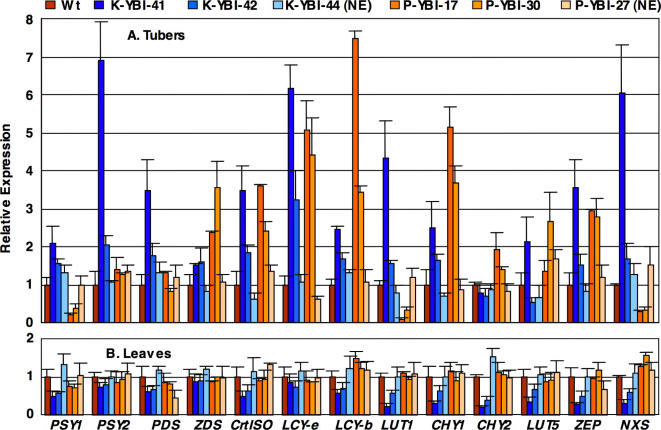
Do the reported genetic manipulations have an influence on endogenous carotenoid gene expression? To answer this question, we measured, via Real Time RT-PCR, the expression of genes for the whole carotenoid pathway in tubers and leaves of the Wt and the transgenic lines. The results (Figure 4) can be summarized as follows: in leaves of the lines constitutively expressing CrtI and CrtY (pK-YBI 41 and 42) a generalized repression is observed, affecting most transcripts, with the exception of PSY2, ZDS, and LCY-e. Although we are not able to determine whether this is a cause or an effect of the decrease in leaf carotenoid content, this is a confirmation that constitutive expression of CrtI and/or CrtY interferes with leaf carotenogenesis; lines expressing CrtI and CrtY in a tuber-specific fashion (pP-YBI 17 and 30) as well as “non-expressor” lines, show instead only minor perturbations in leaf carotenoid transcripts (Figure 4); in tubers of all the lines analyzed, with the exception of the “non-expressor” lines (pK-YBI 44 and pP-YBI 27), a generalized induction of several transcripts is observed (Figure 4); some notable differences are observed between the pK lines (showing intermediate expression of the transgenes in the tubers) and the pP lines (showing very high transgene expression in this tissue, Table 2).
Figure 4. Endogenous carotenoid gene expression.
Transcript levels were measured through Real Time RT-PCR and were first normalized for expression of the housekeeping β-tubulin gene, and then for the expression levels in the Wt. A: tubers. B: leaves. For each construct, two lines with significant carotenoid changes and one “non expressor” line (NE) are shown. The histograms show the average and SE (error bars) of determinations from at least 4 different tubers (or leaves) from 2 different plants. For details see Materials and Methods.
Thus, by introducing a bacterial mini-pathway for carotenoid biosynthesis, it is possible to increase potato tuber carotenoid content to levels much higher than what has been previously described, either in genetically engineered lines or in natural variants. β−carotene increases to the highest levels described to date in any of the 4 major staple crops. Although different conversion rates of β-carotene into retinol have been reported, this conversion can be as high as 3∶1 [18]. Assuming a more conservative, 6∶1 conversion rate, used in human supplementation studies [19], reaching 50% of the vitamin A RDA of 800 µg retinol equivalents would require 250 gms/day (fresh weight) of the “golden” tubers reported in this work. Although no data exist on the bioavailability of β-carotene from potatoes, it should be noticed that, unlike other antioxidants, carotenoids are not degraded or made less bioavailable after cooking. In fact, cooked sweet potatoes, which are naturally rich in β-carotene, have been shown to improve the vitamin A status of children [20].
Differently from “golden” rice and canola [5] [17], in the case of potato all three genes (CrtB, CrtI and CrtY) are necessary for attaining maximal levels of tuber carotenoids. Clearly, the “golden” lines seem to be outliers with respect to the majority of lines produced by the pP-YBI construct (Figure 2), indicating that the presence of the construct by itself is not sufficient to give the “golden tuber” phenotype. However, a χ2 test shows that this construct produces “golden” lines with a frequency (2/9) significantly different from the double construct pP-BI (0/9, P = 1.4×10−4) or the single construct pP-I (0/18, P = 1.3×10−6). Finally, it is essential that all three transgenes are put under tuber-specific promoter control, as the expression in leaves of CrtY and/or CrtI produces a series of alterations in macroscopic phenotypes (Figure 3), biochemical composition (Table 3), and endogenous gene expression (Figure 4).
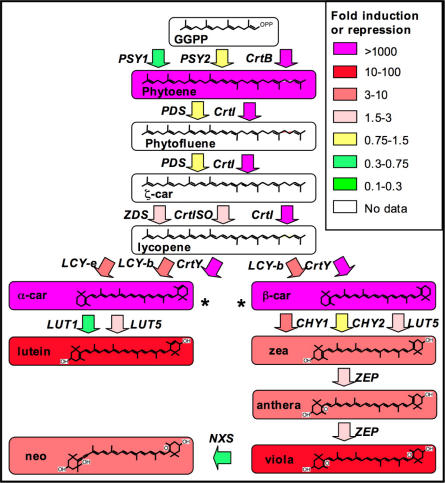
In “golden” tubers, provitamin A carotenoids (α- and β-carotene) increase dramatically (Figure 5) as do a series of downstream compounds (lutein, violaxanthin, neoxanthin). The increase in these compounds is paralleled by the increase in some of the endogenous transcripts involved in their biosynthesis: CrtISO, LCY-e, CHY1, ZEP (Figure 5). Some of the observed perturbations are likely to antagonize the accumulation of provitamin A carotenoids: for instance, the induction of CHY1, LUT5 and ZEP favors the partial conversion of α- and β-carotene into α- and β-xanthophylls [21] [22] [23] [24] [25] [26]. In fact, the levels of these xanthophylls increase, although not as dramatically as α- and β-carotene (Figure 5). These xanthophylls may also have a beneficial nutritional effect, since high dietary intakes of xanthophylls, particularly of lutein, have been associated with a lowered risk for certain cancers and eye degenerative syndromes [27].
Figure 5. Schematic representation of metabolite and gene expression changes in “golden” tubers.
Boxes represent the metabolic intermediates, arrows represent the genes catalyzing the various reactions. Fold induction or repression with respect to the wild-type - averaged over lines P-YBI 17 and 30 - is represented by different color hues (see legend). Asterisks mark Provitamin A carotenoids (α- and β-carotene).
What about the stability of the “golden tuber” trait? Since potato is propagated vegetatively, rather than sexually, we checked the stability through subsequent generations of micropropagated plants. Each transformed line, derived from a single transformation event, has been multiplied in vitro for over two years, and several different harvests have been tested spectrophotometrically and via HPLC, finding only minor variations in carotenoid content (Table 3).
We believe that the carotenoid and provitamin A content of potato can be further improved, with respect to the results shown here: first, we and others have shown that gene silencing can be exploited to re-direct the biosynthetic flux in the pathway, enhancing the accumulation of specific carotenoids such as β-carotene or zeaxanthin [28] [29] [26]; second, the accumulation of phytoene observed in the “golden” tubers indicates that, in spite of the expression of a codon-optimized CrtI gene, phytoene desaturation is still a rate-limiting step; third, Lu et al. [16] have shown that the cauliflower Or gene can be used to increase total carotenoid and β-carotene levels in tubers; since this gene does not appear to increase the capacity for biosynthesis [30], which is the main target of the manipulation reported in this work, it is possible that it will have an additive affect with respect to the constructs reported here. Therefore, a combination of different approaches is very promising for the further improvement of potato carotenoid content, either for nutritional purposes, or for the production of high value-added compounds.
Materials and Methods
Unless indicated differently, molecular biology methods are as described [31]. The Pat1 promoter has been described previously [32] [29], while the Pat2 promoter is a 648-bp fragment from the P24 promoter [33] flanked by Sal I (upstream) and Bam HI (downstream) restriction sites. CrtB and CrtY genes are from Erwinia herbicola [34]. The synthetic CrtI gene [13] and the RbcS transit peptide [15] have been described. The transformation vectors are shown in Figure 1B and they are derivatives of pCAMBIA 1390 [35]. They were constructed as follows:
pK-I: The synthetic TP-CrtI gene was excised from pFun1 [13] using BamHI and ligated into a pCAMBIA1390 derivative containing the CaMV 35S promoter and a kanamycin resistance gene to yield pK-I.
pK-BI: To generate a TP-CrtB fusion, the CrtB gene was amplified from the Erwinia herbicola gene cluster using the following primers: CrtB-Rev: 5′- GATTGAGGCATGCCAATGAGCCA-3′ and CrtB-For: 5′-ACCTACAGGGGTACCTGCGTGA-3′. The obtained fragment was then digested with KpnI, filled-in with the T4-DNA-polymerase, treated with SphI and finally ligated into pTBlue, a pBluescript derivative encoding the RbcS transit peptide [15] and harboring an additional polylinker from pUC18, to yield pTCrtB. The TP-CrtB gene was then isolated through KpnI-digestion and ligated into pAsBB33, a pBluescript derivative harboring the patatin 1 promoter, the ocs terminator and a pUC18 polylinker flanked by two AscI-sites. The TP-CrtB expression cassette was then excised from the obtained plasmid, pTCrtB33, using AscI and ligated into the corresponding site in pK-I to yield pK-BI.
pK-YBI: CrtY was amplified from the Erwinia herbicola gene cluster using the following primers: CrtY-Rev: 5′-GAGAGCGTAGCATGCGGGATCTGA-3′and CrtY-For: 5′-AGCTCGAGGATCCACCAAAGCCTG-3′. The obtained fragment was then digested with SphI/XhoI and ligated into the corresponding sites of pTBlue to create a TP-CrtY fusion gene. The TP-CrtY fragment was then isolated from the generated plasmid, pTCrtY, using BamHI and XbaI and ligated into the corresponding sites of pUCET4 [15] to yield pUCrtY. The generated TP-CrtY expression cassette was then isolated from pUCrtY as a blunt-end fragment through EcoRI/HindIII-digestion and subsequent T4-DNA-polymerase treatment. The fragment was then ligated into EcoRI-digested and blunt-ended pTCrtB33 to yield pTCrtBY33. The fragment encoding the TP-CrtB and TP-CrtY expression cassettes was then isolated from pTCrtBY33 using AscI and ligated into the corresponding site of pK-I to yield pK-YBI
pP-I: The synthetic TP-CrtI gene was excised from pK-I as a BamHI-fragment and ligated into the corresponding site of pPAT1390, a pCAMBIA1390 derivative containing the patatin 2 promoter and a kanamycin resistance gene, to yield pP-I.
pP-BI: The TP-CrtB expression cassette was excised from pTCrtB33 using AscI and ligated into the corresponding site of pP-I to yield pP-BI.
pP-YBI: The patatin 2 promoter was ligated as HindIII/SmaI fragment into XbaI-digested, T4-DNA-polymerase- and HindIII-treated pUCrtY replacing the 35S CaMV promoter to yield pPAT-TCrtY. To isolate the TP-CrtY expression cassette, pPAT-TCrtY was then digested with HindIII/EcoRI and filled-in with the T4-DNA-polymerase. The obtained fragment was then ligated into EcoRI-digested and T4-DNA-polymerase-treated pTCrtB33 to yield pPAT-CrtBY. The fragment encoding the TP-CrtB and TP-CrtY expression cassettes was then isolated from pPAT-CrtBY using AscI and ligated into the corresponding site of pP-I to yield pP-YBI.
Potato (cv Desirée) was transformed and transformants were selected on kanamycin and tested via PCR as described previously [36] [29]. In order to select for independent transformation events, only one regenerant/leaf disk was isolated. PCR primers were: CrtB For: CTG ACC CAC GGT ATT ACG; CrtB Rev: CGT CTT CGC CCG AAT AAC; CrtI For: GCG ACC AGT AGC ATC TAC; CrtI Rev: GTT AGA TGC CAC GGC TTG; CrtY For: CAT TCC ATG AAG ACG ATC TG; CrtY Rev: GCG AAT AGC CAG TGG TAG. Each line was micropropagated [36] and two plantlets were adapted and brought to maturity in the greenhouse [29]. All carotenoid and RT-PCR measurements were conducted on at least 4 different “deep” tubers derived from the 2 plants, to account for plant-to-plant variation and to minimize possible alterations in carotenoid composition/gene expression resulting from light accidentally illuminating the superficial tubers.
RNA isolation, Real Time RT-PCR conditions and primers were described previously [29]. Additional primers used in this work were: Nxs For: CTTGGAGGAGACTTCTTTGGTGA; Nxs Rev: CGGAAGTGGTCCTCCCATAG; Lut5 For: GTCTCAAGCAAGCAACTTCGTG; Lut5 Rev: GATAAAAGGTCCATGTGAGCACTG;
For spectrophotometric quantitation of carotenoids, tuber samples (∼1.5–2.0 g FW) were peeled and homogenized with 2 ml of water with an Ultraturrax homogenizer at full speed until they were completely disrupted; 2 ml of acetone were added and the homogenization was continued for 30”. 2 ml of light Petroleum Ether (PE) were added to each sample and, after vortexing for 2′, the samples were centrifuged at 8000× g for 5′ in a swinging bucket rotor. The 400–500 nm spectrum of the upper phase (PE+carotenoids) was determined against a PE blank. Leaf pigment quantitation was performed as described previously [37].
For HPLC analysis, frozen tubers were peeled, lyophilized, ground to powder and pigments were extracted three times with 2 ml of acetone. In the first extraction, 200 µg tocopherol acetate per sample was added as an internal standard. Combined acetone extracts were dried, lipophilic compounds were resuspended in 2 ml of petroleum ether:diethyl ether (2∶1, v/v) and 1 ml of distilled water. After centrifugation for 5 min at 3000× g the organic phase was recovered and the aqueous phase was extracted for a second time as described above. The combined organic phases were dried and dissolved in 30 µl chloroform. 10 µl were subjected to HPLC analysis with a C30 reversed-phase column (YMC Europe GmbH, Schermbeck, Germany) and a gradient system as described [10]. Carotenoids were identified by their absorption spectra, monitored using a photodiode array detector (PDA 2996; Waters, Eschborn, Germany). Peak areas of individual carotenoids were integrated electronically at the respective λmax. Total carotenoid levels in different lines were normalized using spectrophotometry.
Acknowledgments
GD acknowledges a Ph.D. fellowship from the University of L'Aquila and the supervision of Prof. Laura Spano' for his doctoral work. We thank Giuseppe Puglia and Patrick Schaub for help with spectrophotometric determinations.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The work was supported by the EC projects ProVitA, EU-SOL and DeveloNutri, by the HarvestPlus (www.harvestplus.org) research consortium, and by the Italian Ministry of Research (FIRB project). None of the funding bodies took part in experiment planning, data analysis or in the writing of the paper.
References
- 1.Nesterenko S, Sink KC. Carotenoid profiles of potato breeding lines and selected cultivars. HortScience. 2003;38:1173–1177. [Google Scholar]
- 2.Morris WL, Ducreux L, Griffiths DW, Stewart D, Davies HV, et al. Carotenogenesis during tuber development and storage in potato. J Exp Bot. 2004;55:975–982. doi: 10.1093/jxb/erh121. [DOI] [PubMed] [Google Scholar]
- 3.Griffiths DW, Dale MF, Morris WL, Ramsay G. Effects of season and postharvest storage on the carotenoid content of Solanum phureja potato tubers. J Agric Food Chem. 2007;55:379–385. doi: 10.1021/jf0620822. [DOI] [PubMed] [Google Scholar]
- 4.Giuliano G, Aquilani R, Dharmapuri S. Metabolic engineering of plant carotenoids. Trends Plant Sci. 2000;5:406–409. doi: 10.1016/s1360-1385(00)01749-0. [DOI] [PubMed] [Google Scholar]
- 5.Ye X, Al-Babili S, Kloti A, Zhang J, Lucca P, et al. Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- 6.Shewmaker CK, Sheehy JA, Daley M, Colburn S, Ke DY. Seed-specific overexpression of phytoene synthase: increase in carotenoids and other metabolic effects. Plant J. 1999;20:401–412. doi: 10.1046/j.1365-313x.1999.00611.x. [DOI] [PubMed] [Google Scholar]
- 7.Ducreux LJ, Morris WL, Hedley PE, Shepherd T, Davies HV, et al. Metabolic engineering of high carotenoid potato tubers containing enhanced levels of beta-carotene and lutein. J Exp Bot. 2005;56:81–89. doi: 10.1093/jxb/eri016. [DOI] [PubMed] [Google Scholar]
- 8.Romer S, Fraser PD, Kiano JW, Shipton CA, Misawa N, et al. Elevation of the provitamin A content of transgenic tomato plants. Nat Biotechnol. 2000;18:666–669. doi: 10.1038/76523. [DOI] [PubMed] [Google Scholar]
- 9.Al-Babili S, Beyer P. Golden Rice–five years on the road–five years to go? Trends Plant Sci. 2005;10:565–573. doi: 10.1016/j.tplants.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Hoa TT, Al-Babili S, Schaub P, Potrykus I, Beyer P. Golden Indica and Japonica rice lines amenable to deregulation. Plant Physiol. 2003;133:161–169. doi: 10.1104/pp.103.023457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaub P, Al-Babili S, Drake R, Beyer P. Why is golden rice golden (yellow) instead of red? Plant Physiol. 2005;138:441–450. doi: 10.1104/pp.104.057927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paine JA, Shipton CA, Chaggar S, Howells RM, Kennedy MJ, et al. Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat Biotechnol. 2005;23:482–487. doi: 10.1038/nbt1082. [DOI] [PubMed] [Google Scholar]
- 13.Al-Babili S, Hoa TT, Schaub P. Exploring the potential of the bacterial carotene desaturase CrtI to increase the beta-carotene content in Golden Rice. J Exp Bot. 2006;57:1007–1014. doi: 10.1093/jxb/erj086. [DOI] [PubMed] [Google Scholar]
- 14.Fray RG, Wallace A, Fraser PD, Valero D, Hedden P, et al. Constitutive expression of a fruit phytoene synthase gene in transgenic tomatoes causes dwarfism by redirecting metabolites from the gibberellin pathway. Plant J. 1995;8:693–701. [Google Scholar]
- 15.Misawa N, Yamano S, Linden H, de Felipe MR, Lucas M, et al. Functional expression of the Erwinia uredovora carotenoid biosynthesis gene crtl in transgenic plants showing an increase of beta-carotene biosynthesis activity and resistance to the bleaching herbicide norflurazon. Plant J. 1993;4:833–840. doi: 10.1046/j.1365-313x.1993.04050833.x. [DOI] [PubMed] [Google Scholar]
- 16.Lu S, Van Eck J, Zhou X, Lopez AB, O'Halloran DM, et al. The Cauliflower Or Gene Encodes a DnaJ Cysteine-Rich Domain-Containing Protein That Mediates High-Levels of {beta}-Carotene Accumulation. Plant Cell. 2006;18:3594–3605. doi: 10.1105/tpc.106.046417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravanello MP, Ke D, Alvarez J, Huang B, Shewmaker CK. Coordinate expression of multiple bacterial carotenoid genes in canola leading to altered carotenoid production. Metab Eng. 2003;5:255–263. doi: 10.1016/j.ymben.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Howe JA, Tanumihardjo SA. Carotenoid-biofortified maize maintains adequate vitamin a status in Mongolian gerbils. J Nutr. 2006;136:2562–2567. doi: 10.1093/jn/136.10.2562. [DOI] [PubMed] [Google Scholar]
- 19.West KP, Jr, Katz J, Khatry SK, LeClerq SC, Pradhan EK, et al. Double blind, cluster randomised trial of low dose supplementation with vitamin A or beta carotene on mortality related to pregnancy in Nepal. The NNIPS-2 Study Group. Bmj. 1999;318:570–575. doi: 10.1136/bmj.318.7183.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Jaarsveld PJ, Faber M, Tanumihardjo SA, Nestel P, Lombard CJ, et al. Beta-carotene-rich orange-fleshed sweet potato improves the vitamin A status of primary school children assessed with the modified-relative-dose-response test. Am J Clin Nutr. 2005;81:1080–1087. doi: 10.1093/ajcn/81.5.1080. [DOI] [PubMed] [Google Scholar]
- 21.Duckham SC, Linforth RST, Taylor IB. Abscisic-acid-deficient mutants at the aba gene locus of Arabidopsis thaliana are impaired in the epoxidation of zeaxanthin. Plant Cell Environ. 1991;14:601–606. [Google Scholar]
- 22.Tian L, Magallanes-Lundback M, Musetti V, DellaPenna D. Functional analysis of beta- and epsilon-ring carotenoid hydroxylases in Arabidopsis. Plant Cell. 2003;15:1320–1332. doi: 10.1105/tpc.011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian L, Musetti V, Kim J, Magallanes-Lundback M, DellaPenna D. The Arabidopsis LUT1 locus encodes a member of the cytochrome p450 family that is required for carotenoid epsilon-ring hydroxylation activity. Proc Natl Acad Sci U S A. 2004;101:402–407. doi: 10.1073/pnas.2237237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, DellaPenna D. Defining the primary route for lutein synthesis in plants: the role of Arabidopsis carotenoid beta-ring hydroxylase CYP97A3. Proc Natl Acad Sci U S A. 2006;103:3474–3479. doi: 10.1073/pnas.0511207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiore A, Dall'osto L, Fraser PD, Bassi R, Giuliano G. Elucidation of the beta-carotene hydroxylation pathway in Arabidopsis thaliana. FEBS Lett. 2006;580:4718–4722. doi: 10.1016/j.febslet.2006.07.055. [DOI] [PubMed] [Google Scholar]
- 26.Diretto G, Welsch R, Tavazza R, Mourgues F, Pizzichini D, et al. Silencing of beta-carotene hydroxylase increases total carotenoid and beta-carotene levels in potato tubers. BMC Plant Biol. 2007;7:11. doi: 10.1186/1471-2229-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krinsky NI. Possible biological mechanisms for a protective role of xanthophylls. J Nutr. 2002;132:540S–542S. doi: 10.1093/jn/132.3.540S. [DOI] [PubMed] [Google Scholar]
- 28.Romer S, Lubeck J, Kauder F, Steiger S, Adomat C, et al. Genetic engineering of a zeaxanthin-rich potato by antisense inactivation and co-suppression of carotenoid epoxidation. Metab Eng. 2002;4:263–272. doi: 10.1006/mben.2002.0234. [DOI] [PubMed] [Google Scholar]
- 29.Diretto G, Tavazza R, Welsch R, Pizzichini D, Mourgues F, et al. Metabolic engineering of potato tuber carotenoids through tuber-specific silencing of lycopene epsilon cyclase. BMC Plant Biol. 2006;6:13. doi: 10.1186/1471-2229-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Lu S, Cosman KM, Earle ED, Garvin DF, et al. beta-Carotene accumulation induced by the cauliflower Or gene is not due to an increased capacity of biosynthesis. Phytochemistry. 2006;67:1177–1184. doi: 10.1016/j.phytochem.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual (Second Edition) Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Rocha-Sosa M, Sonnewald U, Frommer W, Stratmann M, Schell J, et al. Both developmental and metabolic signals activate the promoter of a class I patatin gene. Embo J. 1989;8:23–29. doi: 10.1002/j.1460-2075.1989.tb03344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu XY, Rocha-Sosa M, Hummel S, Willmitzer L, Frommer WB. A detailed study of the regulation and evolution of the two classes of patatin genes in Solanum tuberosum L. Plant Mol Biol. 1991;17:1139–1154. doi: 10.1007/BF00028731. [DOI] [PubMed] [Google Scholar]
- 34.To KY, Lai EM, Lee LY, Lin TP, Hung CH, et al. Analysis of the gene cluster encoding carotenoid biosynthesis in Erwinia herbicola Eho13. Microbiology. 1994;140( Pt 2):331–339. doi: 10.1099/13500872-140-2-331. [DOI] [PubMed] [Google Scholar]
- 35.Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 36.Tavazza R, Tavazza M, Ordas RJ, Ancora G, Benvenuto E. Genetic transformation of potato (Solanum tuberosum); an efficient method to obtain transgenic plants. Plant Science. 1988;59:175–181. [Google Scholar]
- 37.Porra RJ, Thompson WA, Kriedermann PE. Determination of accurate extinction coefficients and simultaneous equation for assaying chlorophyll a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta. 1989;975:384–394. [Google Scholar]