Abstract
In pancreatic β-cells, following an acute (within 1 h) increase in glucose concentration, there are rapid changes in the expression of a large subset of proteins. The change in the expression of many of these proteins is mediated by a post-transcriptional mechanism through either increases or decreases in the rate of translation from pre-existing transcripts. These proteins, whose synthesis is rapidly up- or down-regulated in response to glucose, are likely important in mounting the correct response to changes in plasma glucose concentrations. However, the vast majority of these proteins remain unidentified. Therefore, in order to identify these proteins, we analysed changes in the levels of mRNAs associated with polysomes (i.e. actively translating mRNAs) isolated from mouse insulinoma 6 cells incubated at either 0·5 or 20 mM glucose for 1 h. Changes in the levels of polysomal mRNAs in response to glucose were analysed using affymetrix oligonucleotide microarrays (translational profiling). This work revealed that, in response to a change in glucose concentration, the abundance of 313 transcripts associated with polysomes changed by more than 1·5-fold, of which the abundance of 37 changed by more than twofold. The majority of these transcripts encoded proteins associated with metabolism or gene expression. More detailed analysis showed that a number of mRNAs encoding proteins associated with the induction of oxidative stress, including thioredoxin-2 and thioredoxin-interacting protein were rapidly redistributed onto heavier polysomes at high glucose concentration, indicating an increase in their expression. At low glucose concentration, when the general rate of protein synthesis is low, a number of mRNAs encoding integrated stress response proteins, including ATF4 and CHOP10, associate with heavier polysomes, indicating that their expression is up-regulated. In conclusion, translational profiling has revealed that, at either low or at high glucose concentration, β-cells rapidly increase the synthesis of a specific subset of proteins that are likely important in maintaining β-cell integrity and survival during conditions of nutritional stress.
Introduction
The pancreatic β-cell rapidly releases insulin in response to nutrients, such as amino acids or glucose (Campbell et al. 1982). To ensure the immediate replenishment of insulin within the β-cell, there is a rapid increase in proinsulin synthesis (up to 10 to 20-fold within 40 min in response to glucose), which is regulated almost entirely through a post-transcriptional mechanism (Itoh et al. 1978, Itoh & Okamoto 1980). Additionally, there are rapid changes in the synthesis of a large number of other proteins, also mediated through a post-transcriptional mechanism (Itoh & Okamoto 1980, Guest et al. 1989, 1991). The vast majority of these proteins that are rapidly up- or down-regulated in response to glucose remain to be identified, but they are likely to be important in mounting the correct response to changes in plasma glucose concentrations. For example, the biogenesis of the secretory granule requires the co-ordinate synthesis and assembly of a large number of proteins (Guest et al. 1989, 1991). Moreover, inhibiting protein synthesis in islets by the addition of cycloheximide perturbs insulin secretion in response to glucose (Garcia-Barrado et al. 2001). Therefore, alterations in the normal synthesis of these proteins may result in defective storage or secretion of pro/insulin and symptoms associated with type II diabetes.
Although gene expression profiling of β-cells incubated at low versus high glucose concentrations has identified proteins that are transcriptionally regulated by glucose (Webb et al. 2000, 2001, Shalev et al. 2002, Ohsugi et al. 2004), increases in protein expression mediated solely through an increase in the rate of protein synthesis would not have been detected. Therefore, in order to identify proteins regulated by glucose through changes in their rate of protein synthesis, translational profiling of mouse insulinoma 6 (MIN6) cells acutely incubated at either low or high glucose concentration was performed (i.e. microarray analysis was performed on mRNAs associated with polysomes, as an increase in the association of mRNA with polysomes is indicative of an increase in the rate of initiation step of translation and hence an increase in protein expression (Johannes et al. 1999, Mikulits et al. 2000)).
Materials and Methods
Chemicals and materials
Analytical grade biochemicals were purchased from Fisher Scientific or Sigma, unless otherwise specified. Klenow fragment and dNTPs were obtained from Promega. Hybond-N membrane, [α-32P] dCTP redivue tips, RNA guard and probequant G50 columns were obtained from Amersham Biosciences. Foetal calf serum was from Invitrogen.
Cell culture and treatment
MIN6 cells (kindly provided by Prof. Jun-Ichi Miyazaki) were used at approximately 80% confluence between passages 16 and 28. MIN6 cells were grown in Dulbecco's modified Eagle's medium containing 25 mM glucose supplemented with 15% heat-inactivated foetal calf serum, 100 μg/ml streptomycin, 100 units/ml penicillin sulphate and 75 μM β-mercaptoethanol, equilibrated with 5% CO2, 95% air at 37 °C. Prior to treatment, the medium was removed and the cells washed twice in HEPES-balanced Kreb's Ringer bicarbonate buffer (115 mM NaCl, 5 mM KCl, 10 mM NaHCO3, 2·5 mM MgCl2, 2·5 mM CaCl2 and 20 mM HEPES pH 7·4) containing 0·5% BSA (KRB). Cells were then pre-incubated for 1 h at 37 °C in KRB containing 0·5 mM glucose (unless otherwise stated in the figure legends) prior to incubation in KRB containing 0·5 or 20 mM glucose for a further hour (unless otherwise stated in the figure legends). During the final 10 min of treatment, cycloheximide was added to the cells at a concentration of 100 μg/ml to prevent ribosomal run-off.
Insulin ELISA
Insulin secretion assays were performed using the Insulin Mouse ELISA kit (DRG Instruments GmbH, Germany) in accordance with the manufacturer's instructions.
Polysome analysis
After treatment, cells were lysed in polysome buffer (20 mM HEPES pH 7·6, 15 mM MgCl2, 300 mM KCl, 1 mg/ml heparin, 0·1 mg/ml cycloheximide, 1 mM dithiothreitol, 1 μl/ml RNAguard, 1 mM benzamidine-HCl, 0·1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin and 1 μg/ml pepstatin) supplemented with 1% triton. Cell lysates were centrifuged for 10 min at 13 000 g at 4 °C to remove nuclei and cell debris. The supernatants were then layered onto 20–50% sucrose gradients (made in polysome buffer) and centrifuged at 39 000 r.p.m. for 2 h at 4 °C in a Sorvall TH64.1 rotor. The gradients were fractionated using a Teledyne ISCO, NE, USA gradient fractionator that continuously measured absorbance at 254 nm. For northern-blot analysis, RNA was precipitated overnight at −80 °C from each fraction by the addition of three volumes of 8 M guanidine-HCl and four volumes of ethanol. RNA was pelleted by centrifugation, washed in 75% ethanol, dried and resuspended in H2O. For microarray analysis, fractions containing polysomes (more than three ribosomes) were pooled. Three volumes of 8 M guanidine-HCl and 0·1 M sodium acetate was added to 1 volume of the pooled RNA. An equal volume of ethanol was added and RNA was precipitated at −80 °C overnight. RNA was pelleted by centrifugation at 12 000 g for 15 min, washed in 75% ethanol and dried. RNA pellets were then resuspended in H2O and an equal volume of 5 M LiCl was added. RNA was precipitated overnight at −80 °C and then pelleted by centrifugation at 12 000 g for 15 min. RNA pellets were then washed in 75% ethanol, dried under vacuum and resuspended in an appropriate quantity of water.
Microarray analysis
In this study, Affymetrix mouse microarrays mouse expression (MOE) 430A were used. These arrays contain 22 626 probe sets for approximately 14 000 genes. The synthesis of fragmented cRNA from polysomal RNA and the hybridisation of the cRNA to the arrays were carried out by the Microarray facility at the University of Leicester and the MRC gene service at the University of Cambridge. To prepare the labelled targets for hybridisation to the array, polysomal RNA was reverse transcribed using an oligo-dT primer containing a T7 polymerase promoter. T7 polymerase was then used to generate double-stranded cDNA. The double-stranded cDNA was then transcribed in vitro to incorporate biotinylated CTP and UTP. The biotin-labelled cRNA was then fragmented and hybridised to the arrays. The arrays were washed, stained with a streptavidin–phycoerythrin conjugate and finally scanned. To determine changes in the gene expression, the signals from the baseline array (0·5 mM glucose) were then compared with the control array (20 mM glucose). Analysis of the data was carried out using the MicroArray Suite 5 (MAS5, Affymetrix) using the following criteria: (1) probe sets with absent calls in both baseline (0·5 mM glucose) and experimental (20 mM glucose) arrays were excluded; (2) probe sets with no change call between baseline and experimental arrays were excluded and (3) a threshold of 1·5- or 2-fold change in the levels of polysomal mRNAs between low and high glucose concentrations in the two individual experiments was included. Additional analysis was then carried out using Excel.
Northern blotting
RNA samples were heated at 65 °C for 10 min in RNA sample buffer (Sigma). RNA samples were cooled on ice and run on 1% agarose formaldehyde gels. RNA was transferred by capillary action onto Hybond-N membrane (Amersham Biosciences) and fixed. [α-32P]-dCTP radio-labelled cDNA probes were generated using Prime-a-Gene labelling system (Promega). Unincorporated [α-32P]-dCTP was removed by spinning through Probequant G50 columns (Amersham Biosciences). Membranes were pre-hybridised at 65 °C for 1 h in Church Gilbert solution (0·5 M NaHPO4 pH 7·2, 1 mM EDTA, 7% SDS) supplemented with denatured salmon sperm DNA (60 μg/ml) prior to addition of the denatured radio-labelled probe and hybridisation overnight. The membranes were then washed twice for 15 min in 2× SSC, 0·1% SDS at room temperature and once for 15 min in 0·2× SSC, 0·1% SDS at 60 °C. The hybridised cDNA probes were visualised by autoradiography.
SDS-PAGE and western blotting
SDS-PAGE and western blotting were performed as described previously (Gomez et al. 2004). Anti-glutathione peroxidase 4 (GPx4), anti-c-Jun and anti-ATF4 (C-20) were purchased from AbCam Ltd, Cell Signalling Technologies, MA, USA, and Santa Cruz Biotechnology, Inc. Santa Cruz, CA, USA respectively. Anti-thioredoxin interacting protein antibody was purchased from Medical and Biological Laboratories Co. Ltd, Naka-Ku Nagoya, Japan.
Results
Analysis of pancreatic β-cell mRNA translation profiles at both low and high glucose concentrations
Upon an increase in glucose concentration, there are rapid changes in the rate of protein expression through changes in the rate of translation from pre-existing transcripts (Guest et al. 1989, 1991). In order to identify proteins, whose expression is acutely regulated (within 1 h) by glucose through an increase in their rate of protein synthesis, we analysed changes in the levels of mRNAs associated with polysomes (defined here as mRNAs bound to more than three ribosomes) in MIN6 cells, a pancreatic β-cell line that synthesises and secretes insulin in response to physiologically relevant glucose concentrations (Ishihara et al. 1993, Skelly et al. 1996), using affymetrix oligonucleotide microarrays.
MIN6 cells were incubated in KRB supplemented with 0·5 mM glucose for 1 h and then further incubated in KRB supplemented with 0·5 or 20 mM glucose for 1 h, conditions that stimulated a 3·27-fold increase in insulin secretion (Fig. 1). Affymetrix microarray analysis was then used to determine differences in the quantity of polysomal mRNA between low and high glucose concentrations for transcripts from 14 000 genes. This analysis revealed that the levels of 313 transcripts associated with polysomes changed by more than 1·5-fold (see Supplementary Data) between low and high glucose concentration in two separate experiments. The level of 37 of these transcripts changed by more than twofold between low and high glucose concentration in two separate experiments (Table 1). These glucose responsive genes, whose polysomal mRNA levels changed by more than 1·5- and 2-fold between low and high glucose concentrations in two separate experiments, were functionally classified using the National Center for Biotechnology Information (NCBI) databases (www.ncbi.nlm.nih.gov) according to known cellular functions or sequence similarity to genes of known functions (Fig. 2). This classification revealed that the majority of polysomal mRNAs whose levels increased or decreased by 1·5-fold or more in response to increases in glucose concentration are involved in either metabolism or transcription (Fig. 2). This indicates that the acute regulation of metabolism and transcription are likely important for the β-cell to maintain the correct response to changes in glucose concentration. Further analysis revealed that the largest increases in the abundance of polysomal mRNAs encoded proteins associated with oxidative stress, indicating that the expression of these proteins is up-regulated in response to high glucose concentration (Table 2). These included an 8·31-fold glucose-stimulated increase in the amount of polysomal thioredoxin-2 (Trx2) mRNA, a mitochondrial thioredoxin that plays an important role in the thiol anti-oxidative pathway, a 3·78-fold increase in polysomal mRNAs encoding thioredoxin interacting protein (TrxIP) and a 2·62-fold increase in polysomal mRNAs encoding GPx4. The largest decreases in the abundance of polysomal mRNAs were of transcripts encoding proteins associated with the integrated stress response, indicating that the expression of these proteins is maintained/increased at low glucose concentration (Table 2). These included a 4·33-, 3·74- and 3·19-fold decrease in the amount of polysomal mRNA encoding the transcription factors c-Jun, C/EBP homologous protein (CHOP10) and activating transcription factor 4 (ATF4) respectively. Additionally, there was a 2·75-fold decrease in the amount of Tribbles homolog 3, a downstream effector of both CHOP10 and ATF4 (Ohoka et al. 2005, Ord & Ord 2005).
Figure 1.
Glucose stimulated insulin secretion in MIN6 cells. MIN6 cells were pre-incubated in KRB containing 0·5 mM glucose for 1 h followed by incubation in KRB containing 0·5 or 20 mM glucose for a further hour. The amount of insulin secreted into the supernatants was determined by ELISA.
Table 1.
List of polysomal mRNAs whose levels changed by more than twofold in response to an increase in glucose concentration in two independent experiments
| Accession no. | Fold changea | |
|---|---|---|
| Gene description | ||
| Cell signalling | ||
| Plexizn A2 | D86949 | −2·00 |
| Suppressor of cytokine signalling 7 | AK014988 | −2·38 |
| Potassium inwardly-rectifying channel | NM_008426 | −2·47 |
| RTP801 | AK017926 | −2·56 |
| β-Spectrin 2, non-erythrocytic | BQ174069 | −2·64 |
| Tribbles homolog 3 | BC012955 | −2·75 |
| Protein synthesis | ||
| Gene rich cluster, C2f gene (ribosome biogenesis) | NM_0135362 | 2·32 |
| Metabolism | ||
| Thioredoxin 2 (Trx2) | NM_019913 | 8·31 |
| Thioredoxin interacting protein (TrxIP) | NM_023719 | 3·78 |
| Glutathione peroxidase 4 (GPx4) | AF274027 | 2·62 |
| Clone IMAGE:6398163 mRNA (homology to NADH dehygrogenase) | C888802 | 2·14 |
| S-Adenosylmethionine decarboxylase1 | NM_009665 | 2·15 |
| O-Fucoly transferase | BC003494 | 2·30 |
| Mitochondrial carrier homolog 1 | AF192558 | 2·00 |
| Aminolevulinicacid synthase 1 | BC022110 | −2·07 |
| Transcription | ||
| Early growth response 1 (egr1) | NM_007913 | 2·75 |
| FBJ osteosarcoma oncogene (Fos) | AV026617 | 2·23 |
| Amino-terminal enhancer of split | NM_010347 | 2·15 |
| Breakpoint cluster region protein 1 | NM_011793 | 2·07 |
| Core promoter element binding protein | AV025472 | −2·15 |
| RIKEN cDNA2610029D06 gene (zinc finger protein) | BB333454 | −2·23 |
| Inhibitor of DNA binding 3 | NM_008321 | −2·23 |
| Zinc finger protein 93 | NM_009567 | −2·38 |
| Zinc finger protein 54 | NM_011760 | −2·49 |
| Zinc finger protein | BC023090 | −2·59 |
| Zinc finger protein 51 | BC011183 | −2·70 |
| Deltex 2 homolog (Drosophila) | BB518874 | −2·07 |
| Activating transcription factor 4 (ATF4) | AV314773 | −3·19 |
| DNA-damage inducible transcript 3 (CHOP10) | NM_007837 | −3·74 |
| Feminization 1 homolog b (C. elegans) (FEM1b) | NM_010193 | −3·98 |
| Jun-oncogene (c-jun) | BC002081 | −4·33 |
| Secretion | ||
| Rab acceptor 1 | L40934 | 2·39 |
| Rab1B | BC016408 | 2·30 |
| Calpactin | NM_009112 | 2·07 |
| Unclassified/miscellaneous | ||
| Clone IMAGE:5358852, mRNA | BI692833 | 2·15 |
| RIKEN cDNA2700019D07 gene | BM937429 | −2·07 |
| Myosin light chain, alkali, non-muscle | BC026760 | 2·07 |
Average fold change from two independent experiments as calculated by MicroArray Suite 5.
Figure 2.
Functional classification of polysomal mRNAs, whose levels changed by more than 2-fold or more than 1·5-fold in response to an increase in glucose concentration. Polysomal mRNAs whose levels changed by more than 1·5-fold (a) or more than twofold (b) were functionally classified using NCBI databases (www.ncbi.nlm.nih.gov) according to known cellular functions or sequence similarity to genes of known functions. The proportional representation of each classification group was plotted on a pie chart.
Table 2.
List of polysomal mRNAs associated with oxidative stress or the integrated stress response, whose levels changed by more than 1·5-fold in response to an increase in glucose concentration
| Accession no. | Fold changea | |
|---|---|---|
| Gene name | ||
| Oxidative stress | ||
| Thioredoxin 2 (Trx2) | NM_019913 | 8·31 |
| Thioredoxin interacting protein (TrxIP) | NM_023719 | 3·78 |
| Glutathione peroxidase 4 (GPx4) | AF274027 | 2·62 |
| Peroxiredoxin 2 | NM_011563 | 1·62 |
| Integrated stress response | ||
| c-Jun | BC002081 | −4·33 |
| CHOP10 (GADD153) | NM_007837 | −3·74 |
| Activating transcription factor 4 (ATF4) | AV314773 | −3·19 |
| Tribbles homolog 3 (Trb3) | BC012955 | −2·75 |
Average fold change from two independent experiments as calculated by MicroArray Suite 5.
Polysome analysis of candidate anti-oxidative stress and integrated stress response mRNAs
Microarray analysis revealed that, at high glucose concentration, there was an increase in the association of a number of mRNAs with polysomes, encoding proteins implicated in the anti-oxidative pathway (Table 2). To confirm these changes, the sedimentation profiles of Trx2, TrxIP and GPx4 mRNAs were determined in MIN6 cells incubated at either low or high glucose concentration by sucrose sedimentation gradient centrifugation followed by northern-blot analysis (Fig. 2a). Polysome profiles revealed that, upon an increase in glucose concentration from 0·5 to 20 mM, there is a large decrease in the 60S/80S peak and a corresponding increase in the amount of polysomes (Fig. 3ai). This indicates that glucose stimulates the movement of ribosomes onto mRNAs and therefore suggests that glucose stimulates an increase in the global rate of initiation, results similar to those previously reported (Gomez et al. 2004). At low glucose concentration, northern-blot analysis of specific mRNAs across the gradient revealed that the majority of mRNA encoding GPx4, Trx2 and TrxIP are associated with monosomes, disomes or trisomes and only a small proportion is associated with larger polysomes (Fig. 3aii). However, upon an increase in glucose concentration, these mRNAs were recruited onto heavier polysomes, indicative of an increase in the rate of their translation. These results demonstrate that ribosomes are recruited onto TrxIP, Trx2 and GPx4 mRNAs at high glucose and therefore confirm the results obtained by microarray analysis. In order to identify whether these increases in mRNA associated with polysomes were due to changes in the levels of total mRNA, northern-blot analysis was carried out on total mRNA isolated from MIN6 cells incubated at either 0·5 or 20 mM glucose for 1 h (Fig. 3b). Northern-blot analysis revealed that glucose stimulated a 2·8-fold increase in TrxIP mRNA, suggesting that short-term incubation at high glucose concentration led to an increase in either the stability and/or transcription of TrxIP mRNA (Fig. 3b). It has previously been reported that TrxIP is transcriptionally up-regulated in human and mouse islets following a 24-h incubation at high glucose (Shalev et al. 2002, Minn et al. 2005b). Therefore, at high glucose concentration, the expression of this protein is regulated by both a significant increase in its rate of transcription and by the recruitment of its mRNA into polysomes. In contrast, the level of GPx4 was unchanged and the level of Trx2 mRNAs was only marginally increased in response to high glucose concentration (Fig. 3b). Therefore, an increase in the expression of these proteins is likely to be primarily mediated through an increase in the rate of protein synthesis.
Figure 3.
Polysome analysis of mRNAs associated with oxidative stress. MIN6 cells were pre-incubated in KRB containing 0·5 mM glucose for 1 h followed by incubation in KRB containing 0·5 or 20 mM glucose for a further hour. (a) Cells were lysed and polysome analysis was carried out using 20–50% sucrose gradients. The gradients were fractionated from top (fraction 1) to bottom (fraction 20). (a. i) Absorbance of the gradients was measured continuously at 254 nm to give polysome profiles. (a. ii) RNA was isolated from each fraction and run on 1% agarose formaldehyde gel. RNA was transferred onto nylon membrane and probed for the mRNAs indicated. The results presented are representative of three separate experiments. (b) Cells were lysed and total RNA isolated. The RNA was then run on 1% agarose formaldehyde gel, transferred onto nylon membrane and probed for specific mRNAs as shown. The fold changes observed in the northern blot between low and high glucose concentrations were quantified with ImageJ. The results presented in this figure are representative of three separate experiments.
In contrast, microarray analysis showed that the expression of a number of polysomal mRNAs associated with the integrated stress response was lower at high glucose concentration (see Table 2). To confirm these changes, the sedimentation profiles of CHOP10, ATF4 and c-Jun mRNAs were determined in MIN6 cells incubated at either low or high glucose by sucrose sedimentation gradient centrifugation followed by northern-blot analysis (Fig. 3a). Paradoxically, at low glucose concentration, CHOP10, ATF4 and c-Jun mRNAs were all found to co-sediment with heavy polysomes, indicating that these mRNAs are likely to be actively engaged in protein synthesis at low glucose (Fig. 3aii). Upon an increase in glucose concentration, there was a significant shift of CHOP10 and ATF4 mRNA off polysomes and into monosomes, indicative of a decrease in their expression (Fig. 3aii). Therefore, at low glucose concentration, the expressions of CHOP10 and ATF4 are likely up-regulated through an increase in their rate of translation. In contrast, c-Jun mRNA remained associated with polysomes but the level of c-Jun mRNA decreased dramatically. To ascertain whether the expression of these proteins was also regulated through changes in mRNA abundance, total RNA was isolated from MIN6 cells that were incubated at low or high glucose concentration and the mRNA levels of ATF4, CHOP10 and c-Jun were determined by northern-blot analysis. Northern blots showed that CHOP10 mRNA levels were 1·7-fold higher at low glucose compared with high glucose, indicating that glucose acutely regulates the expression of this protein by both transcriptional and translational mechanisms. ATF4 mRNA levels were only marginally increased at low glucose. Therefore, the increase in the expression of ATF4 at low glucose is likely primarily mediated through an increase in the rate of protein synthesis. In contrast, the level of c-Jun mRNA decreased dramatically between low and high glucose (8·9-fold decrease) and therefore the expression of this protein is likely primarily regulated by glucose through changes in mRNA abundance.
Glucose regulates the protein levels of anti-oxidative stress and integrated stress response proteins
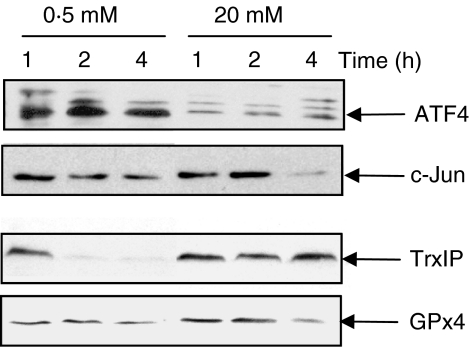
To ascertain whether the changes in the association of mRNA with polysomes identified by microarray analysis correlated with changes in protein expression, MIN6 cells were incubated at 0·5 or 20 mM glucose for up to 4 h and the expression levels of a number of candidate proteins determined by western blotting (Fig. 4). The protein expression levels of ATF4 were much higher at low glucose concentration than at high glucose concentration throughout the period of the experiment. The level of c-Jun protein significantly decreased after 4 h incubation at high glucose concentration, whereas the levels of c-Jun protein were maintained for up to 4 h at low glucose concentration. These results provide evidence that the rate of c-Jun and ATF4 protein synthesis is higher at low glucose concentration than at high glucose concentration. These results are consistent with our microarray data demonstrating that ATF4 and c-Jun mRNAs are recruited onto polysomes at low glucose concentration. In contrast, TrxIP protein levels were rapidly decreased at low glucose (within 1–2 h) but maintained at high glucose concentration. These results are also consistent with our microarray results demonstrating that TRxIP is recruited onto polysomes at high glucose concentration. The protein levels of GPx4 also increased after 1–2 h at high glucose concentration, again consistent with our microarray data.
Figure 4.
Glucose regulated protein expression. MIN6 cells were incubated in KRB at either 0·5 or 20 mM glucose for the times indicated. Samples of cell lysates were run on a SDS-polyacrylamide gel followed by western blotting using antisera against ATF4, c-Jun, TrxIP and GPx4.
Supplementary data
A list of polysomal mRNAs whose levels changed by more than 1.5-fold in response to an increase in glucose concentration is presented (http://joe.endocrinology-journals.org/cgi/content/full/192/1/DC1). Polysomal mRNAs whose levels were shown to increase or decrease by more than 1.5 fold in response to 1 h glucose stimulation in two separate experiments are listed. Analysis was carried out using MAS5.
Discussion
Translational profiling of the pancreatic β-cell line MIN6 revealed the identity of a large number of mRNAs that are rapidly redistributed on or off polysomes in response to changes in glucose concentration, indicative of changes in their rates of synthesis. Functional analysis of these glucose-regulated genes showed that 34 and 46% of mRNAs, whose abundance on polysomes changed by more than 1·5- and 2-fold respectively were associated with gene expression (Fig. 2).
In response to high glucose concentration, the general rate of protein synthesis increases, primarily mediated through an increase in the rate of translation initiation (Gomez et al. 2004). However, paradoxically, the association of a number of transcripts with polysomes increased in response to a decrease in glucose concentration, indicating that translation from these transcripts is up-regulated at low glucose concentration. These included transcripts that encode b-zip transcription factors, ATF4, CHOP10 and c-Jun, whose expression has previously been shown to be up-regulated in response to an increase in the phosphorylation of eIF2α, as part of the integrated stress response (Harding et al. 2003). Indeed, we have previously demonstrated that eIF2α is phosphorylated in response to low glucose concentration in MIN6 cells (Gomez et al. 2004). The amount of total ATF4 mRNA is slightly higher at low glucose compared with high glucose concentration, suggesting that glucose deprivation also stimulates the transcription of ATF4 or increases the stability of ATF4 mRNA. However, the redistribution of ATF4 mRNA on polysomes between high and low glucose indicate that its expression is primarily regulated at the post-transcriptional level through an increase in the rate of protein synthesis. Indeed, the mechanism by which ATF4 is up-regulated when general translation is repressed through the phosphorylation of eIF2α is known (Lu et al. 2004, Vattem & Wek 2004). Essentially, ATF4 expression is regulated by short open reading frames (uORFs) within its 5′ untranslated region (5′UTR) by a mechanism analogous to that which regulates general control non depressible 4 (GCN4) expression in yeast (Lu et al. 2004, Vattem & Wek 2004). Briefly, GCN4 mRNA contains four upstream open reading frames (uORFs) in its 5′UTR. Under non-stressed conditions, when eIF2α is unphosphorylated and the availability of the translational ternary complex is high, the ribosome initiates at the 5′ proximal uORF. Upon termination of uORF1, the ribosome remains associated with the mRNA and, due to the availability of ternary complex, is able to reinitiate at inhibitory ORF 2, 3 and 4 which overlaps the coding region of GCN4, thereby bypassing the authentic initiation codon. In stressed conditions, when eIF2α is phosphorylated and the availability of the translational ternary complex is low, there is a delay in reinitiation that allows the ribosome to bypass the inhibitory uORFs and initiate at the authentic initiation codon, resulting in the expression of GCN4 (Hinnebusch 1994).
At low glucose concentration, the mRNA encoding CHOP10 (GADD153), a downstream target of ATF4 (Harding et al. 2000), and c-jun were also found to be associated with polysomes, indicating that the expression of these proteins is up-regulated at low glucose concentration. Northern-blot analysis of total RNA indicates that there are acute changes in the abundance of these mRNA in response to glucose, indicating that both changes in the mRNA stability/rates of transcription are clearly important in regulating their expression. Microarray analysis of MIN6 cells incubated at low or high glucose concentration for 24 h showed that CHOP expression was increased at low glucose concentration (Webb et al. 2000, 2001). The mechanism by which the synthesis of CHOP10 and c-Jun is maintained at low glucose concentration is unknown. However, it is likely that the rapid up-regulation of these integrated stress response proteins are necessary for the β-cell to respond to stress induced by glucose deprivation.
At high glucose concentration, a number of mRNAs that play an important role in the regulation of oxidative stress, including key members of the thioredoxin and glutathione anti-oxidative systems moved onto heavy polysomes, indicative of an increase in their rates of translation and therefore protein expression. These included the mitochondrial thioredoxin Trx2, GPx4 and peroxiredoxin-2 (Table 2). Changes in abundance of mRNAs on polysomes were independent of changes in transcript levels. Therefore, any changes in the expression of these proteins in response to an acute increase in glucose concentration are mediated at the post-transcriptional level via an increase in the rate of translation from pre-existing mRNA.
The β-cell is particularly vulnerable to oxidative stress, probably due to its low anti-oxidative enzyme activity, which makes it difficult to inactivate reactive oxygen species (Grankvist et al. 1981, Lenzen et al. 1996, Tiedge et al. 1997). This weakness of the β-cell to combat oxidative stress is thought to be an important factor in the development of diabetes, as it can lead to β-cell dysfunction and apoptosis, a major form of β-cell loss in both type I and type II diabetes. As it has previously been demonstrated that the over-expression of thioredoxin-1 protects mice from streptozotocin-induced diabetes (Hotta et al. 1998) and the over-expression of glutathione peroxidase protects β-cells from oxidative stress (Robertson et al. 2005), it is likely that the up-regulation of Trx2; GPx4 and peroxiredoxin-2 are important in protecting against hyperglycaemia-induced oxidative stress. Paradoxically, microarray analysis indicated that, in response to high glucose concentration, there was also an increase in the expression of TrxIP, a protein that binds to the active site of thioredoxin-1 and inhibits its activity. This increase in expression is primarily mediated through an increase in transcript number, as glucose stimulated a large increase in TrxIP mRNA abundance (Fig. 3). Indeed, it has previously been demonstrated, in both islets and pancreatic β-cells, that glucose stimulates the transcription of TrxIP (Minn et al. 2005a). Up-regulating TrxIP results in the inhibition of thioredoxin's reactive oxygen species scavenging function, resulting in increased cellular oxidative stress. This may be an important mechanism by which hyperglycaemia induces oxidative stress and subsequent pancreatic β-cells apoptosis (Schulze et al. 2004). Indeed, over-expression of TrxIP induces apoptosis in islets (Minn et al. 2005a). Interestingly, glutathione peroxidase and thioredoxin have also been implicated in regulating glucose-stimulated insulin secretion mediated by increases in NADPH (Ivarsson et al. 2005).
This study does not provide a comprehensive list of proteins, whose expression is acutely up- or down-regulated in response to glucose. Indeed, it is important to note that the expression of proteins, whose mRNA abundance does not appear to change on polysomes could still be acutely up- or down-regulated by glucose. However, changes in the abundance of mRNAs on polysomes in response to glucose detected in this report likely parallels the changes in the expression levels of the proteins they encode. Indeed, this study provides evidence that hyperglycaemia up-regulates a number of proteins responsible for regulating the redox status of the β-cell. Deregulation of the redox state of the cell by glucose is likely to play an important role in the development of β-cell dysfunction and type II diabetes. Additionally, this study revealed that the integrated stress response is activated at low glucose concentration, which is an important mechanism for alleviating ER stress and therefore increasing cell survival (Harding et al. 2003).
In conclusion, this study has identified a number of proteins that are acutely regulated by glucose. Further analysis and characterisation of the function of these proteins in response to changing glucose concentration is likely to provide a better understanding of the processes important in the development of diabetes.
Acknowledgements
I C G was supported by a Biotechnology and Biological Sciences Research Council case studentship. E G was supported the Wellcome Trust. C M is supported by an MRC studentship. The authors declare that there is no conflict of interest that would prejudice the impartiality of this scientific work.
Supplementary Material
References
- Campbell I.L, Hellquist L.N, Taylor K.W. Insulin biosynthesis and its regulation. Clinical Science. 1982;62:449–455. doi: 10.1042/cs0620449. [DOI] [PubMed] [Google Scholar]
- Garcia-Barrado M.J, Ravier M.A, Rolland J.F, Gilon P, Nenquin M, Henquin J.C. Inhibition of protein synthesis sequentially impairs distinct steps of stimulus-secretion coupling in pancreatic beta cells. Endocrinology. 2001;142:299–307. doi: 10.1210/endo.142.1.7910. [DOI] [PubMed] [Google Scholar]
- Gomez E, Powell M.L, Greenman I.C, Herbert T.P. Glucose-stimulated protein synthesis in pancreatic β-cells parallels an increase in the availability of the translational ternary complex (eIF2-GTP.Met-tRNAi) and the dephosphorylation of eIF2α. Journal of Biological Chemistry. 2004;279:53937–53946. doi: 10.1074/jbc.M408682200. [DOI] [PubMed] [Google Scholar]
- Grankvist K, Marklund S.L, Taljedal I.B. CuZn-superoxide dismutase, Mn-superoxide dismutase, catalase and glutathione peroxidase in pancreatic islets and other tissues in the mouse. Biochemical Journal. 1981;199:393–398. doi: 10.1042/bj1990393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest P.C, Rhodes C.J, Hutton J.C. Regulation of the biosynthesis of insulin-secretory-granule proteins. Co-ordinate translational control is exerted on some, but not all, granule matrix constituents. Biochemical Journal. 1989;257:431–437. doi: 10.1042/bj2570431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest P.C, Bailyes E.M, Rutherford N.G, Hutton J.C. Insulin secretory granule biogenesis. Co-ordinate regulation of the biosynthesis of the majority of constituent proteins. Biochemical Journal. 1991;274:73–78. doi: 10.1042/bj2740073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Molecular Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Harding H.P, Zhang Y, Zeng H, Novoa I, Lu P.D, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Molecular Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. Translational control of GCN4: an in vivo barometer of initiation-factor activity. Trends in Biochemical Sciences. 1994;19:409–414. doi: 10.1016/0968-0004(94)90089-2. [DOI] [PubMed] [Google Scholar]
- Hotta M, Tashiro F, Ikegami H, Niwa H, Ogihara T, Yodoi J, Miyazaki J. Pancreatic beta cell-specific expression of thioredoxin, an antioxidative and antiapoptotic protein, prevents autoimmune and streptozotocin-induced diabetes. Journal of Experimental Medicine. 1998;188:1445–1451. doi: 10.1084/jem.188.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H, Asano T, Tsukuda K, Katagiri H, Inukai K, Anai M, Kikuchi M, Yazaki Y, Miyazaki J.I, Oka Y. Pancreatic beta cell line MIN6 exhibits characteristics of glucose metabolism and glucose-stimulated insulin secretion similar to those of normal islets. Diabetologia. 1993;36:1139–1145. doi: 10.1007/BF00401058. [DOI] [PubMed] [Google Scholar]
- Itoh N, Okamoto H. Translational control of proinsulin synthesis by glucose. Nature. 1980;283:100–102. doi: 10.1038/283100a0. [DOI] [PubMed] [Google Scholar]
- Itoh N, Sei T, Nose K, Okamoto H. Glucose stimulation of the proinsulin synthesis in isolated pancreatic islets without increasing amount of proinsulin mRNA. FEBS Letters. 1978;93:343–347. doi: 10.1016/0014-5793(78)81136-3. [DOI] [PubMed] [Google Scholar]
- Ivarsson R, Quintens R, Dejonghe S, Tsukamoto K, in't Veld P, Renstrom E, Schuit F.C. Redox control of exocytosis: regulatory role of NADPH, thioredoxin, and glutaredoxin. Diabetes. 2005;54:2132–2142. doi: 10.2337/diabetes.54.7.2132. [DOI] [PubMed] [Google Scholar]
- Johannes G, Carter M.S, Eisen M.B, Brown P.O, Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. PNAS. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzen S, Drinkgern J, Tiedge M. Low antioxidant enzyme gene expression in pancreatic islets compared with various other mouse tissues. Free Radical Biology and Medicine. 1996;20:463–466. doi: 10.1016/0891-5849(96)02051-5. [DOI] [PubMed] [Google Scholar]
- Lu P.D, Harding H.P, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. Journal of Cell Biology. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulits W, Pradet-Balade B, Habermann B, Beug H, Garcia-Sanz J.A, Mullner E.W. Isolation of translationally controlled mRNAs by differential screening. FASEB Journal. 2000;14:1641–1652. doi: 10.1096/fj.14.11.1641. [DOI] [PubMed] [Google Scholar]
- Minn A.H, Hafele C, Shalev A. Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces beta-cell apoptosis. Endocrinology. 2005;146:2397–2405. doi: 10.1210/en.2004-1378. [DOI] [PubMed] [Google Scholar]
- Minn A.H, Pise-Masison C.A, Radonovich M, Brady J.N, Wang P, Kendziorski C, Shalev A. Gene expression profiling in INS-1 cells overexpressing thioredoxin-interacting protein. Biochemical and Biophysical Research Communications. 2005;336:770–778. doi: 10.1016/j.bbrc.2005.08.161. [DOI] [PubMed] [Google Scholar]
- Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO Journal. 2005;24:1243–1255. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsugi M, Cras-Meneur C, Zhou Y, Warren W, Bernal-Mizrachi E, Permutt M.A. Glucose and insulin treatment of insulinoma cells results in transcriptional regulation of a common set of genes. Diabetes. 2004;53:1496–1508. doi: 10.2337/diabetes.53.6.1496. [DOI] [PubMed] [Google Scholar]
- Ord D, Ord T. Characterization of human NIPK (TRB3, SKIP3) gene activation in stressful conditions. Biochemical and Biophysical Research Communications. 2005;330:210–218. doi: 10.1016/j.bbrc.2005.02.149. [DOI] [PubMed] [Google Scholar]
- Robertson R.P, Tanaka Y, Takahashi H, Tran P.O, Harmon J.S. Prevention of oxidative stress by adenoviral overexpression of glutathione-related enzymes in pancreatic islets. Annals of the New York Academy of Sciences. 2005;1043:513–520. doi: 10.1196/annals.1333.058. [DOI] [PubMed] [Google Scholar]
- Schulze P.C, Yoshioka J, Takahashi T, He Z, King G.L, Lee R.T. Hyperglycemia promotes oxidative stress through inhibition of thioredoxin function by thioredoxin-interacting protein. Journal of Biological Chemistry. 2004;279:30369–30374. doi: 10.1074/jbc.M400549200. [DOI] [PubMed] [Google Scholar]
- Shalev A, Pise-Masison C.A, Radonovich M, Hoffmann S.C, Hirshberg B, Brady J.N, Harlan D.M. Oligonucleotide microarray analysis of intact human pancreatic islets: identification of glucose-responsive genes and a highly regulated TGFbeta signaling pathway. Endocrinology. 2002;143:3695–3698. doi: 10.1210/en.2002-220564. [DOI] [PubMed] [Google Scholar]
- Skelly R.H, Schuppin G.T, Ishihara H, Oka Y, Rhodes C.J. Glucose-regulated translational control of proinsulin biosynthesis with that of the proinsulin endopeptidases PC2 and PC3 in the insulin-producing MIN6 cell line. Diabetes. 1996;45:37–43. doi: 10.2337/diab.45.1.37. [DOI] [PubMed] [Google Scholar]
- Tiedge M, Lortz S, Drinkgern J, Lenzen S. Relation between antioxidant enzyme gene expression and antioxidative defense status of insulin-producing cells. Diabetes. 1997;46:1733–1742. doi: 10.2337/diab.46.11.1733. [DOI] [PubMed] [Google Scholar]
- Vattem K.M, Wek R.C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. PNAS. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb G.C, Akbar M.S, Zhao C, Steiner D.F. Expression profiling of pancreatic beta cells: glucose regulation of secretory and metabolic pathway genes. PNAS. 2000;97:5773–5778. doi: 10.1073/pnas.100126597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb G.C, Akbar M.S, Zhao C, Steiner D.F. Expression profiling of pancreatic beta cells: glucose regulation of secretory and metabolic pathway genes. Diabetes. 2001;50(Suppl 1):S135–S136. doi: 10.2337/diabetes.50.2007.s135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.