Abstract
This paper presents Salmonella data from animals, feedstuffs and feed mills in Sweden between 1993 and 1997. During that period, 555 isolates were recorded from animals, representing 87 serotypes. Of those, 30 serotypes were found in animals in Sweden for the first time. The majority of all isolates from animals were S. Typhimurium (n = 91), followed by S. Dublin (n = 82). There were 115 isolates from cattle, 21 from broilers, 56 from layers and 18 from swine. The majority of these isolates were from outbreaks, although some were isolated at the surveillance at slaughterhouses. The number of isolates from the feed industry was similar to that of the previous 5-year period. Most of those findings were from dust and scrapings from feed mills, in accordance with the HACCP programme in the feed control programme. It can be concluded that the occurrence of Salmonella in animals and in the feed production in Sweden remained favourable during 1993–97.
Keywords: animal, cattle, feed, feed production, isolate, poultry, swine, Salmonella, Sweden
Introduction
Salmonellosis is one of the most common food borne zoonoses reported world-wide [5,18]. However, in Sweden the prevalence of Salmonella in food producing animals is low [7,3,17]. This is most likely due to the Salmonella control programme that started in 1961 with the aim to keep meat- and egg producing animals free from Salmonella. When Sweden joined the European Union (EU) in 1995, surveillance of Salmonella in cattle, pigs and poultry at slaughter was included in the control programme [2].
Any finding of Salmonella from animals or the feed production, regardless of serotype, is notifiable to the Swedish Board of Agriculture (SBA). At least one isolate from each finding of Salmonella in animals, feed or environmental sampling from feed mills has to be sent to the National Veterinary Institute (SVA) for confirmation and serotyping. This is performed according to the methods described by [9]. From each notifiable incident of Salmonella one isolate has to be tested for antibiotic resistance at the SVA. Apart from this, isolates of S. Typhimurium and S. Enteritidis are phage typed at the Swedish Institute for Infectious Disease Control (SMI). In January 1996, the phage typing system was changed from the Lilleengen to the Colindale system [1,19].
The reporting of Salmonella has resulted in a series of articles by the SVA and the SBA with results presented from 1949 and onward [16,13,10,8,6,14,12,4,11]. The aim of the present study is to summarise Salmonella data from animals and the feed production in Sweden between 1993 and 1997.
Materials and methods
The results presented in this study were based on information collected at the SVA and the SBA. If several isolates of the same sero- and phage type were obtained from the same animal or from the same epidemiological unit (i.e. cattle farm, pig farm, kennel, water in reptile terrariums) only the first isolate was included (i.e. primary isolate). If Salmonella was re-isolated after an animal, herd or flock had been cleared from the infection, this isolate was also included. If more than one sero- or phage type was isolated from each individual or epidemiological unit, each serotype was included. Furthermore, isolates from autopsies, sanitary slaughter and lymph nodes collected at the surveillance at the slaughterhouses, were also included even if Salmonella could not be re-isolated at follow-up sampling at the farms. From feed production, all primary isolates were included.
Results and discussion
Salmonella isolated from animals
In total, 555 isolates were recorded from animals during the present study period. Between 1989 and 1992, 598 isolates were recorded. However, comparisons of results between the different study periods must be made with caution as sampling strategy and surveillance may have differed [16,13,10,8,6,14,12,4,11].
In the present study, 78% of the isolates were S. Subspecies I, followed by S. Subspecies III (13%) and II (5%) (Table 1). The number of isolates of S. Subspecies I were fewer compared with results from the previous studies, which most likely is due to the decrease in number of isolates from cattle (Fig. 1). In all, but one, of the previous reports, cattle have been the most common animal specie from which Salmonella was isolated. However, in the present report, reptiles predominated. Most of those isolates were S. Subspecies II, III and IV. An explanation for this may be the increased import of reptiles since March 1996 when the Swedish import regulations were harmonised with the EU regulations. It is likely that the increase in number of reptiles led to increased sampling of this animal specie.
Table 1.
The number of isolates of the various subspecies of Salmonella enterica in animals in Sweden during 1968–97.
| Salmonella enterica subspecies | 1968–72 | 1973–77 | 1978–82 | 1983–87 | 1988–92 | 1993–97 |
| Subspecies I (Subsp. enterica) | 1721 | 1077 | 1231 | 720 | 524 | 435 |
| Subspecies II (Subsp. salamae) | 10 | 2 | 4 | 6 | 11 | 29 |
| Subspecies IIIa & IIIb (Subsp. arizonae & diarizonae) | 14 | 19 | 14 | 13 | 59 | 73 |
| Subspecies IV (Subsp. houtenae) | 1 | 2 | 1 | 3 | 4 | 15 |
| Not typed or typable | 6 | 16 | 18 | 4 | 3 | |
| Total | 1752 | 1116 | 1268 | 746 | 598 | 555 |
Figure 1.
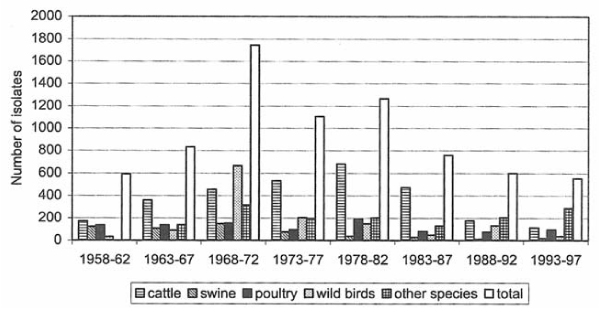
The number of recorded Salmonella isolates from various animal species 1958–97 in Sweden.
During 1993–97, 87 different serotypes were identified from animals (Table 2), which is the largest number ever recorded. Of those, 30 were found in animals in Sweden for the first time. The most common serotype was S. Typhimurium (n = 91), followed by S. Dublin (n = 82), which is in accordance with results from the previous study periods. Table 2 presents the distributions of serotypes during the study period. Two different phage typing system were used for S. Typhimurium in 1993–97. Up to 1995, the Lilleengen system was used, followed by the Colindale system introduced in 1996 (Table 3).
Table 2.
The distribution of serotypes of Salmonella isolated from animals between 1993 and 1997.
| Serotype | Last isolation before 1993 | 1993 | 1994 | 1995 | 1996 | 1997 | Total |
| S. Abony | 1978 | 2 | 2 | 4 | |||
| S. Adamstua | 1969 | 1 | 1 | ||||
| S. Adelaide | 1988 | 4 | 4 | ||||
| S. Afula | 1 | 1 | |||||
| S. Agona | 1992 | 1 | 2 | 1 | 1 | 5 | |
| S. Agoueve | 1 | 1 | |||||
| S. Anatum | 1992 | 1 | 1 | 2 | 4 | ||
| S. Arechavaleta | 1 | 1 | |||||
| S. Bardo | 1991 | 2 | 2 | 4 | |||
| S. Bassa | 1 | 1 | |||||
| S. Bignona | 1 | 1 | |||||
| S. Bissau | 1 | 1 | |||||
| S. Bovismorbificans | 1992 | 1 | 2 | 3 | |||
| S. Braendrup | 1987 | 1 | 1 | 2 | |||
| S. Bredeney | 1991 | 1 | 1 | ||||
| S. Burgas | 2 | 2 | |||||
| S. California | 1984 | 1 | 1 | ||||
| S. Chailey | 1 | 1 | |||||
| S. Chester | 1982 | 1 | 1 | ||||
| S. Cubana | 1983 | 1 | 2 | 1 | 4 | ||
| S. Derby | 1991 | 4 | 4 | ||||
| S. Dublin | 1992 | 22 | 24 | 16 | 13 | 7 | 82 |
| S. Dusseldorf | 1992 | 1 | 1 | 2 | |||
| S. Durban | 1988 | 1 | 1 | ||||
| S. Enteritidis | 1992 | 2 | 1 | 4 | 12 | 1 | 20 |
| S. Finkenwerden | 1984 | 1 | 1 | ||||
| S. Fluntern | 1 | 1 | |||||
| S. Fresno | 1978 | 4 | 4 | ||||
| S. Giza | 1 | 1 | |||||
| S. Hadar | 1991 | 1 | 1 | 2 | |||
| S. Havana | 1984 | 1 | 1 | ||||
| S. Idikan | 1 | 1 | 2 | ||||
| S. Indiana | 1982 | 1 | 1 | ||||
| S. Infantis | 1992 | 3 | 6 | 1 | 1 | 11 | |
| S. Ituri | 1 | 1 | |||||
| S. Java | 1979 | 2 | 1 | 1 | 4 | ||
| S. Kingston | 1 | 1 | |||||
| S. Korovi | 1 | 1 | |||||
| S. Kottbus | 1981 | 1 | 1 | ||||
| S. Koumra | 1 | 1 | |||||
| S. Legon | 1 | 1 | |||||
| S. Lexington | 1991 | 1 | 1 | ||||
| S. Limete | 1 | 1 | |||||
| S. Lindern | 1969 | 1 | 1 | ||||
| S. Linguere | 1 | 1 | |||||
| S. Livingstone | 1992 | 4 | 19 | 7 | 5 | 5 | 40 |
| S. Lomita | 1 | 1 | |||||
| S. Mbandaka | 1992 | 1 | 1 | 3 | 1 | 6 | |
| S. Montevideo | 1989 | 1 | 3 | 2 | 6 | ||
| S. Mowanjum | 1 | 1 | |||||
| S. Muenchen | 1989 | 1 | 2 | 3 | |||
| S. Muenster | 1979 | 3 | 3 | ||||
| S. Nanga | 1 | 1 | |||||
| S. New York | 1 | 1 | |||||
| S. Newport | 1988 | 1 | 1 | 2 | 6 | 6 | 16 |
| S. Nima | 1 | 1 | |||||
| S. Ohio | 1992 | 1 | 1 | ||||
| S. Oranienburg | 1990 | 1 | 3 | 4 | |||
| S. Oslo | 1982 | 2 | 2 | ||||
| S. Panama | 1980 | 1 | 1 | ||||
| S. Plymouth | 3 | 3 | |||||
| S. Poona | 1987 | 2 | 1 | 3 | |||
| S. Potengi | 1 | 1 | |||||
| S. Ramatgan | 1973 | 2 | 2 | ||||
| S. Reading | 1968 | 1 | 1 | ||||
| S. Rissen | 1992 | 2 | 2 | ||||
| S. Rubinslaw | 1976 | 4 | 4 | ||||
| S. Ruiru | 1972 | 1 | 1 | ||||
| S. San-diego | 1988 | 1 | 4 | 1 | 6 | ||
| S. Saintpaul | 1989 | 1 | 1 | ||||
| S. Sao | 1 | 1 | |||||
| S. Saphra | 2 | 2 | |||||
| S. Schwabach | 1 | 1 | |||||
| S. Schwarzengrund | 1992 | 1 | 1 | ||||
| S. Sendai | 1968 | 1 | 1 | ||||
| S. Senftenberg | 1990 | 3 | 1 | 4 | |||
| S. Shanghai | 1981 | 1 | 1 | ||||
| S. Sheffield | 1991 | 1 | 1 | ||||
| S. Stanley | 1990 | 2 | 2 | ||||
| S. Tennessee | 1992 | 1 | 1 | 1 | 3 | ||
| S. Thompson | 1987 | 1 | 1 | 2 | |||
| S. Tshiongwe | 1984 | 1 | 1 | ||||
| S. Typhimurium | 1992 | 26 | 12 | 18 | 21 | 14 | 91 |
| S. Welikade | 1988 | 1 | 1 | ||||
| S. Widemarsh | 2 | 1 | 3 | ||||
| S. Windermere | 1986 | 1 | 1 | ||||
| S. Virginia | 1 | 1 | 2 | ||||
| S. Species | 2 | 1 | 3 | ||||
| S. Subspecies I | 1992 | 4 | 5 | 3 | 4 | 16 | |
| S. Subspecies II | 1992 | 12 | 2 | 11 | 4 | 29 | |
| S. Subspecies III | 1992 | 2 | 2 | 4 | |||
| S. Subspecies IIIa | 8 | 10 | 6 | 6 | 5 | 35 | |
| S. Subspecies IIIb | 1 | 7 | 15 | 11 | 34 | ||
| S. Subspecies IV | 1992 | 3 | 1 | 8 | 1 | 2 | 15 |
| Total | 94 | 108 | 123 | 146 | 84 | 555 | |
I = enterica, II = salamae, III = arizone or diarizonae, IV = houtenae
Table 3.
Phage typing of Salmonella Typhimurium strains isolated from animals 1993–97.
| Lillengen system 1993–95 | ||||||||||||||||
| Species/phage type | 1 | 8 | 9 | 12 | 15 | 22 | LNT | LNST | uk1 | Total | ||||||
| Broilers | 2 | 2 | ||||||||||||||
| Cats | 1 | 1 | 2 | |||||||||||||
| Cattle | 2 | 1 | 2 | 1 | 1 | 1 | 8 | |||||||||
| Dogs | 1 | 1 | 4 | 1 | 7 | |||||||||||
| Horses | 3 | 2 | 1 | 6 | ||||||||||||
| Layers | 1 | 1 | 1 | 1 | 4 | |||||||||||
| Lizards & snakes | 1 | 1 | 1 | 3 | ||||||||||||
| Other domestic fowls2 | 1 | 1 | 2 | |||||||||||||
| Swine | 2 | 2 | ||||||||||||||
| Wild birds | 5 | 2 | 9 | 4 | 20 | |||||||||||
| Total | 14 | 3 | 14 | 4 | 2 | 1 | 1 | 13 | 4 | 56 | ||||||
| Colindale system 1996–97 | ||||||||||||||||
| Species/phage type | 1 | 2 | 12 | 40 | 41 | 85 | 104 | 120 | 129 | 170 | 195 | 196 | LNT | LNST | u k1 | Total |
| Cats | 2 | 2 | ||||||||||||||
| Cattle | 2 | 1 | 3 | 2 | 2 | 1 | 11 | |||||||||
| Dogs | 1 | 1 | 1 | 3 | ||||||||||||
| Horses | 1 | 1 | ||||||||||||||
| Other domestic fowls2 | 1 | 2 | 1 | 4 | ||||||||||||
| Swine | 2 | 1 | 1 | 1 | 1 | 1 | 7 | |||||||||
| Wild birds | 2 | 5 | 7 | |||||||||||||
| Total | 2 | 2 | 1 | 10 | 3 | 1 | 3 | 1 | 1 | 3 | 3 | 1 | 1 | 2 | 1 | 35 |
1Unknown, 2duck, goose, turkey
The change of phage typing makes comparisons with previous results difficult.
Salmonella isolated from cattle
In cattle, 115 isolates representing 9 different serotypes were found (Tables 4, 5, 6, 7, 8). In Fig. 2 it is shown that the annual number of isolates from cattle has decreased during the last ten years. Seventy-eight isolates emanated from infected herds. The remaining isolates were collected at autopsies, sanitary slaughter and surveillance at slaughterhouses when Salmonella could not be re-isolated at follow-up sampling at the farms. The most commonly isolated serotype in cattle was S. Dublin (n = 76), followed by S. Typhimurium (n = 21), which is similar to findings presented in the previous reports. There were three S. Typhimurium DT 104 isolates phage typed in the Colindale system, one in 1996 and two in 1997 (Table 3). The isolate from 1996 could not be re-isolated in the herd of origin. Apart from this, there was one isolate of S. Typhimurium phage typed as LNT from 1995 that was retyped as DT 104. The strains were resistant to ampicillin, chloramphenicol, streptomycin, sulphonamides and tetracycline.
Table 4.
Salmonella serotypes isolated from animals in Sweden in 1993.
| Broilers | Cattle | Cats | Dogs | Horses | Layers | Lizards & snakes | Swine | Turkey | Turtles | Wild birds | Total | |
| S. Agona | 1 | 1 | ||||||||||
| S. Anatum | 1 | 1 | ||||||||||
| S. Braenderup | 1 | 1 | ||||||||||
| S. Dublin | 22 | 22 | ||||||||||
| S. Dusseldorf | 1 | 1 | ||||||||||
| S. Enteritidis | 1 | 1 | 2 | |||||||||
| S. Livingstone | 4 | 4 | ||||||||||
| S. Lomita | 1 | 1 | ||||||||||
| S. Mbandaka | 1 | 1 | ||||||||||
| S. Newport | 1 | 1 | ||||||||||
| S. Ruiru | 1 | 1 | ||||||||||
| S. Typhimurium | 1 | 2 | 2 | 3 | 3 | 2 | 13 | 26 | ||||
| S. Species1 | 1 | 1 | 2 | |||||||||
| S. Subspecies I | 1 | 3 | 4 | |||||||||
| S. Subspecies II | 1 | 6 | 5 | 12 | ||||||||
| S. Subspecies III | 2 | 2 | ||||||||||
| S. Subspecies IIIa | 8 | 8 | ||||||||||
| S. Subspeceis IIIb | 1 | 1 | ||||||||||
| S. Subspecies IV | 3 | 3 | ||||||||||
| Total | 1 | 25 | 3 | 3 | 4 | 8 | 27 | 2 | 1 | 7 | 13 | 94 |
1Not typable
Table 5.
Salmonella serotypes isolated from animals in Sweden in 1994.
| Broilers | Cage birds | Cattle | Dogs | Layers | Lizards & snakes | Sheep | Various animals1 | Wild birds | Zoo animals2 | Total | |
| S. Agona | 1 | 1 | 2 | ||||||||
| S. Anatum | 1 | 1 | |||||||||
| S. California | 1 | 1 | |||||||||
| S. Dublin | 24 | 24 | |||||||||
| S. Enteritidis | 1 | 1 | |||||||||
| S. Idikan | 1 | 1 | |||||||||
| S. Indiana | 1 | 1 | |||||||||
| S. Infantis | 1 | 2 | 3 | ||||||||
| S. Java | 1 | 1 | 2 | ||||||||
| S. Kingston | 1 | 1 | |||||||||
| S. Korovi | 1 | 1 | |||||||||
| S. Koumra | 1 | 1 | |||||||||
| S. Lexington | 1 | 1 | |||||||||
| S. Livingstone | 2 | 16 | 1 | 19 | |||||||
| S. Mbandaka | 1 | 1 | |||||||||
| S. Montevideo | 1 | 1 | |||||||||
| S. Newport | 1 | 1 | |||||||||
| S. Ohio | 1 | 1 | |||||||||
| S. Oranienburg | 1 | 1 | |||||||||
| S. Reading | 1 | 1 | |||||||||
| S. Rissen | 1 | 1 | 2 | ||||||||
| S. Rubinslaw | 4 | 4 | |||||||||
| S. San-diego | 1 | 1 | |||||||||
| S. Sao | 1 | 1 | |||||||||
| S. Senftenberg | 1 | 2 | 3 | ||||||||
| S. Tennessee | 1 | 1 | |||||||||
| S. Thompson | 1 | 1 | |||||||||
| S. Typhimurium | 1 | 4 | 1 | 2 | 1 | 3 | 12 | ||||
| S. Subspecies I | 1 | 1 | 2 | 1 | 5 | ||||||
| S. Subspecies III | 1 | 1 | 2 | ||||||||
| S. Subspeceies IIIa | 9 | 1 | 10 | ||||||||
| S. Subspecies IV | 1 | 1 | |||||||||
| Total | 10 | 2 | 33 | 7 | 27 | 17 | 1 | 2 | 3 | 6 | 108 |
11 Mouse (Indiana), 1 polecat (Livingstone)
24 Crocodiles (Java, Reading, Subsp III, Subsp IIIa), 2 marsupials (Newport, Thompson)
Table 6.
Salmonella serotypes isolated from animals in Sweden in 1995.
| Broilers | Cattle | Dogs | Horses | Layers | Lizards & snakes | Other domestic fowls1 | Swine | Turtles | Various animals2 | Wild birds | Zoo animals3 | Total | |
| S. Abony | 2 | 2 | |||||||||||
| S. Adelaide | 4 | 4 | |||||||||||
| S. Agona | 1 | 1 | |||||||||||
| S. Agoueve | 1 | 1 | |||||||||||
| S. Anatum | 2 | 2 | |||||||||||
| S. Bardo | 2 | 2 | |||||||||||
| S. Bovismorbificans | 1 | 1 | |||||||||||
| S. Burgas | 2 | 2 | |||||||||||
| S. Chester | 1 | 1 | |||||||||||
| S. Cubana | 1 | 1 | |||||||||||
| S. Dublin | 12 | 4 | 16 | ||||||||||
| S. Durban | 1 | 1 | |||||||||||
| S. Enteritidis | 1 | 2 | 1 | 4 | |||||||||
| S. Fluntern | 1 | 1 | |||||||||||
| S. Giza | 1 | 1 | |||||||||||
| S. Infantis | 1 | 3 | 1 | 1 | 6 | ||||||||
| S. Limete | 1 | 1 | |||||||||||
| S. Livingstone | 3 | 1 | 3 | 7 | |||||||||
| S. Mbandaka | 2 | 1 | 3 | ||||||||||
| S. Montevideo | 1 | 2 | 3 | ||||||||||
| S. Muenchen | 1 | 1 | |||||||||||
| S. New York | 1 | 1 | |||||||||||
| S. Newport | 2 | 2 | |||||||||||
| S. Nima | 1 | 1 | |||||||||||
| S. Oslo | 2 | 2 | |||||||||||
| S. San-diego | 4 | 4 | |||||||||||
| S. Saphra | 2 | 2 | |||||||||||
| S. Schwarzengrund | 1 | 1 | |||||||||||
| S. Senftenberg | 1 | 1 | |||||||||||
| S. Tennessee | 1 | 1 | |||||||||||
| S. Thompson | 1 | 1 | |||||||||||
| S. Tshiongwe | 1 | 1 | |||||||||||
| S. Typhimurium | 4 | 1 | 3 | 2 | 1 | 2 | 5 | 18 | |||||
| S. Virginia | 1 | 1 | |||||||||||
| S. Subspecies I | 1 | 1 | 1 | 3 | |||||||||
| S. Subspecies II | 2 | 2 | |||||||||||
| S. Subspeceies IIIa | 6 | 6 | |||||||||||
| S. Subspeceis IIIb | 6 | 1 | 7 | ||||||||||
| S. Subspecies IV | 8 | 8 | |||||||||||
| Total | 5 | 21 | 4 | 4 | 10 | 43 | 5 | 3 | 9 | 5 | 8 | 6 | 123 |
11 Pheasant (Agona), 2 ostriches (Anatum), 1 turkey (Mbandaka), 1 goose (Typhimurium)
21 Bear (Nima), 4 mink (Dublin)
31 Cayman (Enteritidis), 1 frog (Limete), 1 marsipual (Typhimurium), 1 monkey (Subsp IIIb)
Table 7.
Salmonella serotypes isolated from animals in Sweden in 1996.
| Broilers | Cage bird | Cat | Cattle | Dogs | Horses | Layers | Lizards & snakes | Other domestic fowls1 | Swine | Turtles | Various animals2 | Wild birds | Total | |
| S. Abony | 2 | 2 | ||||||||||||
| S. Adamstua | 1 | 1 | ||||||||||||
| S. Afula | 1 | 1 | ||||||||||||
| S. Agona | 1 | 1 | ||||||||||||
| S. Bardo | 2 | 2 | ||||||||||||
| S. Bignona | 1 | 1 | ||||||||||||
| S. Bissau | 1 | 1 | ||||||||||||
| S. Bovismorbificans | 2 | 2 | ||||||||||||
| S. Braenderup | 1 | 1 | ||||||||||||
| S. Cubana | 2 | 2 | ||||||||||||
| S. Derby | 1 | 1 | 2 | 4 | ||||||||||
| S. Dublin | 11 | 1 | 1 | 13 | ||||||||||
| S. Enteritidis | 1 | 5 | 5 | 1 | 12 | |||||||||
| S. Finkenwerden | 1 | 1 | ||||||||||||
| S. Fresno | 4 | 4 | ||||||||||||
| S. Hadar | 1 | 1 | ||||||||||||
| S. Havana | 1 | 1 | ||||||||||||
| S. Idikan | 1 | 1 | ||||||||||||
| S. Infantis | 1 | 1 | ||||||||||||
| S. Java | 1 | 1 | ||||||||||||
| S. Lindern | 1 | 1 | ||||||||||||
| S. Linguere | 1 | 1 | ||||||||||||
| S. Livingstone | 1 | 4 | 5 | |||||||||||
| S. Mbandaka | 1 | 1 | ||||||||||||
| S. Muenchen | 1 | 1 | 2 | |||||||||||
| S. Muenster | 3 | 3 | ||||||||||||
| S. Newport | 1 | 5 | 6 | |||||||||||
| S. Plymouth | 3 | 3 | ||||||||||||
| S. Poona | 1 | 1 | 2 | |||||||||||
| S. Ramatgan | 2 | 2 | ||||||||||||
| S. Sandiego | 1 | 1 | ||||||||||||
| S. Saint-paul | 1 | 1 | ||||||||||||
| S. Shanghai | 1 | 1 | ||||||||||||
| S. Stanley | 2 | 2 | ||||||||||||
| S. Typhimurium | 2 | 8 | 2 | 1 | 1 | 3 | 4 | 21 | ||||||
| S. Widermarsh | 2 | 2 | ||||||||||||
| S. Windermere | 1 | 1 | ||||||||||||
| S. Virginia | 1 | 1 | ||||||||||||
| S. Subspecies I | 1 | 1 | 2 | 4 | ||||||||||
| S. Subspecies II | 4 | 7 | 11 | |||||||||||
| S. Subspecies IIIa | 5 | 1 | 6 | |||||||||||
| S. Subspecies IIIb | 1 | 14 | 15 | |||||||||||
| S. Subspecies IV | 1 | 1 | ||||||||||||
| Total | 3 | 5 | 2 | 25 | 3 | 3 | 6 | 43 | 11 | 6 | 30 | 4 | 5 | 146 |
11 pheasant (Agona), 7 geese (4 Enteritidis, 3 Muenster), 2 ostrich (Enteritidis, Idikan), 1 duck (Typhimurium)
23 hedghogs (1 Enteritidis, 2 Widermarsh), 1 fox (Dublin)
Table 8.
Salmonella serotypes isolated from animals in Sweden in 1997.
| Broilers | Cage birds | Cattle | Dogs | Horses | Layers | Lizards & snakes | Other domestic fowls1 | Swine | Turtles | Zoo animals2 | Wild birds | Total | |
| S. Arechavaleta | 1 | 1 | |||||||||||
| S. Bassa | 1 | 1 | |||||||||||
| S. Bredeney | 1 | 1 | |||||||||||
| S. Chailey | 1 | 1 | |||||||||||
| S. Cubana | 1 | 1 | |||||||||||
| S. Dublin | 7 | 7 | |||||||||||
| S. Dusseldorf | 1 | 1 | |||||||||||
| S. Enteritidis | 1 | 1 | |||||||||||
| S. Hadar | 1 | 1 | |||||||||||
| S. Infantis | 1 | 1 | |||||||||||
| S. Ituri | 1 | 1 | |||||||||||
| S. Java | 1 | 1 | |||||||||||
| S. Kottbus | 1 | 1 | |||||||||||
| S. Legon | 1 | 1 | |||||||||||
| S. Livingstone | 1 | 4 | 5 | ||||||||||
| S. Montevideo | 1 | 1 | 2 | ||||||||||
| S. Mowanjum | 1 | 1 | |||||||||||
| S. Nanga | 1 | 1 | |||||||||||
| S. Newport | 1 | 5 | 6 | ||||||||||
| S. Oranienburg | 2 | 1 | 3 | ||||||||||
| S. Panama | 1 | 1 | |||||||||||
| S. Poona | 1 | 1 | |||||||||||
| S. Potengi | 1 | 1 | |||||||||||
| S. Schwabach | 1 | 1 | |||||||||||
| S. Sendai | 1 | 1 | |||||||||||
| S. Sheffield | 1 | 1 | |||||||||||
| S. Tennessee | 1 | 1 | |||||||||||
| S. Typhimurium | 3 | 1 | 3 | 4 | 3 | 14 | |||||||
| S. Welikade | 1 | 1 | |||||||||||
| S. Widemarsh | 1 | 1 | |||||||||||
| S. Species3 | 1 | 1 | |||||||||||
| S. Subspecies II | 3 | 1 | 4 | ||||||||||
| S. Subspeceies IIIa | 1 | 3 | 1 | 5 | |||||||||
| S. Subspeceis IIIb | 1 | 10 | 11 | ||||||||||
| S. Subspecies IV | 2 | 2 | |||||||||||
| Total | 2 | 1 | 11 | 3 | 1 | 5 | 35 | 4 | 7 | 8 | 1 | 6 | 84 |
11 Duck (Enteritidis), 3 geese (Typhimurium)
21 Monkey
3Not typable
Figure 2.
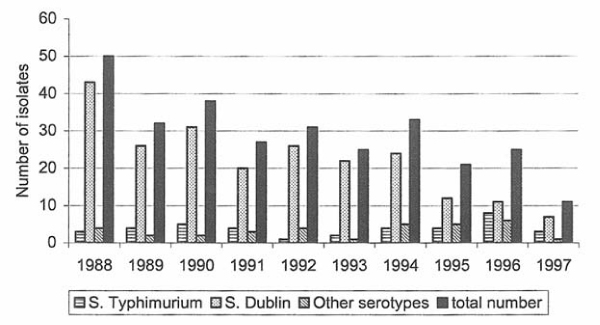
Recorded number of Salmonella isolates from cattle during 1988–97.
Salmonella isolated from swine
In swine, 18 isolates were reported representing 8 serotypes (Tables 4, 5, 6, 7, 8). The number of isolates varied from 2 to 7 per year (Fig. 3). Eight of the isolates were from infected herds and the remaining were collected at sanitary slaughter or at the slaughterhouse surveillance, when Salmonella could not be re-isolated at follow-up sampling on the farm. The most common serotype was S. Typhimurium (n = 9), followed by S. Derby and S. Infantis (n = 2, respectively).
Figure 3.
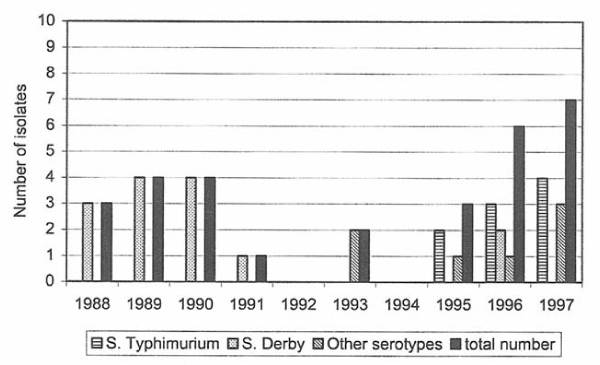
Recorded number of Salmonella isolates from swine during 1988–97.
Salmonella isolated from fowl
Twenty-one isolates were from broilers and 56 from layers. An explanation for the higher number of isolates from layers may be that the Salmonella control programme was implemented in the broiler production earlier than in the egg production. Salmonella Livingstone was the most commonly isolated serotype and seven of the isolates (33%) were from broilers and 31 (55%) from layers (Tables 4, 5, 6, 7, 8). During the last years, the annual number of isolates from layers, broilers and other domesticated fowls has decreased (Fig. 4). In 1994 there were 16 S. Livingstone isolates from layers and it was suspected that this was due to contamination of feed mills, which subsequently may have spread to poultry by the feed. Another more plausible explanation is that the industry led Salmonella control programme that was implemented among laying hens in 1991 became mandatory in 1994 and thereby increased the chance of finding Salmonella through intensified sampling.
Figure 4.
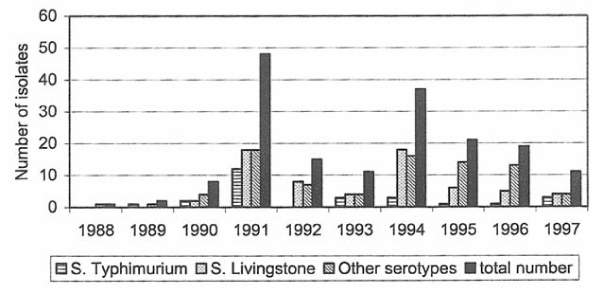
Recorded number of almonella isolates from layers, broilers and other domesticated fowl during 1988–97.
There were 19 Salmonella strains isolated from domestic fowl other than broilers and layers, such as geese (n = 10), ostriches (n = 4), turkeys (n = 3) and ducks (n = 2). Most isolates were S. Enteritidis (n = 6) and S. Typhimurium (n = 4; Tables 4, 5, 6, 7, 8). Furthermore, 35 isolates were from wild birds, of which the majority were S. Typhimurium (n = 28; Tables 4, 5, 6, 7, 8). The most common phage type in small passerine birds in the Colindale system was DT 40 (n = 5; Table 3).
Salmonella in companion animals
In dogs, there were 20 isolates of which S. Typhimurium was the most commonly isolated serotype (n = 8; Tables 4, 5, 6, 7, 8). There were 13 serotypes recorded in total. From cats there were one S. Braenderup and 4 S. Typhimurium isolates. Furthermore, 8 isolates were obtained from cage birds, and 4 of these came from the same zoological garden and were of S. Subspecies IIIa.
Salmonella in zoo, wild and farmed animals
Thirteen isolates were found in 6 species of zoo animals (Tables 4, 5, 6, 7, 8). Of those were 4 isolates from crocodiles and marsupials, respectively. Apart from this, there were 11 isolates from various other animal species, possibly farmed as well as wild.
Salmonella in reptiles
Out of the 555 isolates from animals, 165 (30%) were from snakes and lizards (Tables 4, 5, 6, 7, 8). The number of isolates from these 2 species showed a great increase compared with results from 1988–92 (n = 47). The majority of isolates were S. Subspecies III (n = 60), IV (n = 15) and II (n = 13). From turtles, there were 54 isolates, compared with 14 in 1988–92. Fifteen of the 54 isolates were S. Subspecies II. The increase in number of isolates from reptiles was probably the result of an increased sampling due to increased import when the Swedish import regulations were harmonised with the EU regulations in 1996.
Salmonella in feed production
The monitoring of commercial feed production follows the principles of HACCP based on identified risk factors [15]. The system was initiated in 1991 and has been in operation for more than 12 years. A thorough monitoring of the production line has proved to be an effective means to prevent Salmonella contamination of feed for food producing animals. The samples investigated were from critical control points in the production line mostly consisting of dust samples and scrapings.
A minimum of five samples was taken each week at feed mills producing poultry feed. Other mills producing feed for animal production collected samples from 2 critical control points. The total number of Salmonella findings from the critical control points was 464 (Table 9). The dominating serotypes were S. Livingstone (n = 62), S. Senftenberg (n = 37), S. Cubana (n = 35) and S. Mbandaka (n = 30). More prevalent serotypes in animal production such as S. Typhimurium, S. Enteritidis or S. Dublin were rarely detected in the feed production. However, S. Livingstone was frequently isolated from layers in 1994 and was found to be the most common serotype in feed production. Not previously reported serotypes from feed production was detected during the time period. A number of subtyping investigations were carried out using PFGE (pulse- field gel electrophoresis) to study the possible transmission of Salmonella from feedstuffs to animals.
Table 9.
Salmonella isolated from feedingstuffs and feed processing plants in Sweden 1993–97.
| Raw materials | |||||||
| Serotypes | Vegetable origin | Animal origin | Raw materials, unspecified | Compound feed | Dust and scrapings from feed mills | Pet chews | Unspecified |
| S. Aarhus | 1 | ||||||
| S. Aberdeen | 1 | ||||||
| S. Abony | 1 | 1 | |||||
| S. Agona | 15 | 1 | 2 | 29 | 8 | ||
| S. Alachua | 7 | ||||||
| S. Albany | 1 | 1 | |||||
| S. Altona | 1 | ||||||
| S. Amsterdam | 3 | 1 | 5 | 1 | |||
| S. Anatum | 15 | 1 | 14 | 13 | |||
| S. Babelsberg | 1 | ||||||
| S. Barteilly | 1 | ||||||
| S. Be | 1 | ||||||
| S. Bere | 4 | 4 | 1 | ||||
| S. Bergen | 2 | ||||||
| S. Bonariensis | 1 | ||||||
| S. Brandenburg | 4 | 1 | 1 | ||||
| S. Bredeney | 1 | 1 | 1 | 3 | 1 | ||
| S. California | 2 | 1 | 1 | ||||
| S. Cerro | 1 | 1 | 2 | 1 | |||
| S. Chester | 1 | ||||||
| S. Chincol | 1 | ||||||
| S. Colorado | 1 | ||||||
| S. Corvallis | 1 | ||||||
| S. Cubana | 15 | 1 | 35 | 22 | |||
| S. Derby | 2 | 3 | 3 | 4 | |||
| S. Dublin | 1 | 2 | 1 | ||||
| S. Dusseldorf | 5 | 1 | |||||
| S. Ealing | 1 | ||||||
| S. Emek | 2 | 1 | 3 | 2 | |||
| S. Enteritidis | 4 | 1 | 1 | ||||
| S. Florida | 1 | ||||||
| S. Freemantle (S.II) | 1 | ||||||
| S. Freetown | 1 | 1 | |||||
| S. Gatuni | 1 | ||||||
| S. Give | 1 | ||||||
| S. Gloucester | 1 | ||||||
| S. Hadar | 3 | ||||||
| S. Havana | 8 | 4 | 13 | 1 | 7 | ||
| S. Heidelberg | 1 | 1 | 2 | ||||
| S. Hofit | 1 | ||||||
| S. Idikan | 1 | ||||||
| S. Infantis | 1 | 1 | 3 | 8 | |||
| S. Irachau | 1 | ||||||
| S. Irumu | 1 | ||||||
| S. Isangi | 1 | 2 | 1 | ||||
| S. Java | 1 | ||||||
| S. Jerusalem | 1 | 1 | 1 | ||||
| S. Kainji | 1 | ||||||
| S. Kapemba | 1 | ||||||
| S. Kentucky | 4 | 2 | 4 | 1 | |||
| S. Kibi | 1 | ||||||
| S. Kingston | 3 | 3 | |||||
| S. Kinondoni | 1 | ||||||
| S. Konstanz | 1 | ||||||
| S. Kortrijk | 1 | ||||||
| S. Lamberhurs | 1 | ||||||
| S. Leno | 1 | ||||||
| S. Lexington | 6 | 8 | 1 | ||||
| S. Liverpool | 3 | 3 | 1 | ||||
| S. Livingstone | 7 | 4 | 3 | 62 | 18 | ||
| S. Llandoff | 1 | 6 | 1 | ||||
| S. London | 2 | ||||||
| S. Madelia | 1 | ||||||
| S. Mandoff | 1 | ||||||
| S. Mbandaka | 20 | 7 | 30 | 16 | |||
| S. Meleagridis | 1 | 1 | 2 | 3 | |||
| S. Montevideo | 2 | 5 | 3 | 12 | 8 | ||
| S. Muenchen | 1 | ||||||
| S. Muenster | 1 | 2 | |||||
| S. Newport | 2 | ||||||
| S. Norwich | 1 | ||||||
| S. Ohio | 3 | 11 | 3 | ||||
| S. Ohlstedt | 1 | ||||||
| S. Oranienburg | 1 | 3 | |||||
| S. Orion | 3 | 1 | |||||
| S. Oslo | 1 | ||||||
| S. Othmarschen | 1 | ||||||
| S. Ouakam | 4 | 1 | |||||
| S. Pakistan | 1 | ||||||
| S. Poona | 3 | 3 | |||||
| S. Rideau | 1 | ||||||
| S. Rissen | 3 | 1 | |||||
| S. Ruiru | 2 | 1 | 2 | ||||
| S. Saint Paul | 1 | ||||||
| S. Saloniki | 1 | ||||||
| S. Sambre | 1 | 1 | |||||
| S. San Diego | 1 | ||||||
| S. Schleissheim | 1 | 1 | |||||
| S. Schoeneberg | 1 | ||||||
| S. Schwarzengrund | 1 | ||||||
| S. Seegefeld | 1 | ||||||
| S. Senftenberg | 23 | 6 | 1 | 37 | 20 | ||
| S. Slade | 1 | ||||||
| S. Taksony | 1 | ||||||
| S. Tees | 1 | ||||||
| S. Tennessee | 14 | 2 | 2 | 2 | 15 | 6 | |
| S. Typhimurium | 1 | 21 | 9 | ||||
| S. Vejle | 1 | ||||||
| S. Virchow | 1 | ||||||
| S. Warragul | 1 | ||||||
| S. Weltevreden | 1 | ||||||
| S. Westhampton | 1 | ||||||
| S. Westphalia | 1 | ||||||
| S. Worthington | 6 | 4 | |||||
| S. Subspecies | 5 | 2 | 8 | 6 | |||
| S. Subspecies I | 21 | 1 | 14 | 30 | 17 | ||
| S. Subspecies II | 2 | ||||||
| S. Subspecies IIIa | 1 | ||||||
| Total | 194 | 28 | 48 | 12 | 464 | 3 | 211 |
Only Salmonella negative raw materials may be used in feed production, hence contaminated raw materials must undergo decontamination before use in the production of animal feed. In raw materials of vegetable origin 194 Salmonella isolates were recorded. The most frequently occurring serotypes were S. Senftenberg (n = 23), S. Mbandaka (n = 20), S. Agona, S. Anatum, S. Cubana (each n = 15) and S. Subspecies I (n = 21). The most frequently imported feed raw materials in which Salmonella was isolated were soybean meal, maize and rapeseed products. The most common serotype in raw materials of animal origin was S. Senftenberg (n = 6) with a total of 28 positive samples. Few findings were made in finished feed including pet food (n = 12).
During 1993–97, the total number of positive samples from the feed sector was 749, which was similar to the previous 5-year period. In the current period the greater part of isolates were from critical control points in the feed production, whereas in the last report over half of the isolates emanated from raw materials of animal origin. There were considerably more findings in raw materials of vegetable origin in the present period compared to the one previous, which clearly indicates that feed raw materials are important carriers of Salmonella infection. It seems reasonable to assume that the surveillance programme with sampling according to HACCP principles has largely been successful in finding Salmonella before it reaches the finished feed product.
Conclusion
From the data presented in this study, it can be concluded that Salmonella in animals and in the feed production remained favourable in Sweden during 1993–97. It may be suggested that this was due to the Salmonella control programme in food producing animals and the testing in the feed production according to the HACCP principles. The final aim is to keep the whole chain of food production free from Salmonella contamination.
References
- Anderson ES, Ward LR, Saxe DEMJ, Sa DEJDH. Bacteriophage-typing designations of Salmonella typhimurium. J of Hygiene. 1977;78:297–300. doi: 10.1017/s0022172400056187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous . The Swedish Salmonella control programmes for live animals, eggs and meats. National Veterinary Institute, Swedish Board of Agriculture, National Food Administration; 16 January 1995. [Google Scholar]
- Anonymous . In: Zoonoses in Sweden up to and including 1999. Wahlström H, editor. National Veterinary Institute, National Food Administration, Swedish Board of Agriculture, Swedish Institute for Infectious Disease Control; 2001. [Google Scholar]
- Eld K, Gunnarsson A, Holmberg T, Hurvel B, Wierup M. Salmonella isolated from animals and feedstuffs in Sweden during 1983–1987. Acta Vet Scand. 1991;32:261–277. doi: 10.1186/BF03546988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TM, Motarjemi Y, Miyagawa S, Käferstein FK, Stohr K. Foodborne salmonellosis. World Health Stat Q. 1997;50:81–89. [PubMed] [Google Scholar]
- Gunnarsson A, Hurvell B, Nordblom B, Rutqvist L, Thal E. Salmonella isolated from animals and feedstuffs in Sweden over the period 1968–1972. Nord Vet Med. 1974;26:499–517. [PubMed] [Google Scholar]
- Hopp P, Wahlström H, Hirn J. A common Salmonella Control Programme in Finland, Norway and Sweden. Acta Vet Scand Suppl. 1999;91:45–49. [PubMed] [Google Scholar]
- Hurvell B, Lagerquist U, Rutqvist L, Thal E. Salmonella isolated from animals and feed stuffs in Sweden during 1963–1967. Nord Vet Med. 1969;21:289–305. [Google Scholar]
- Kaufmann F. Serological diagnosis of Salmonella species. Kaufmann-White-Schema. Munksgaard, Copenhagen, Denmark; 1972. [Google Scholar]
- Karlsson K-A, Rutqvist L, Thal E. Salmonella isolated from animals and animal feeds in Sweden during 1958–1962. Nord Vet Med. 1963;15:833–850. [Google Scholar]
- Malmqvis M, Jacobsson K-G, Häggblom P, Cerenius F, Sjöland L, Gunnarsson A. Salmonella isolated from animals and feedstuffs in Sweden during 1988–1992. Acta Vet Scand. 1995;36:21–39. doi: 10.1186/BF03547700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mårtensson L, Holmberg T, Hurvell B, Rutqvist L, Sandstedt K, Wierup M. Salmonella isolated from animals and feedstuffs in Sweden during 1978–1982. Nord Vet Med. 1984;36:371–393. [PubMed] [Google Scholar]
- Rutqvist L, Thal E. Salmonella isolated from animals and animal products in Sweden during 1956–1957. Nord Vet Med. 1958;10:234–244. [Google Scholar]
- Sandstedt K, Gunnarsson A, Hurvell B, Nordblom B, Rutqvist L, Söderlind O. Salmonella isolated from animals and feedstuffs in Sweden during 1973–1977. Nord Vet Med. 1980;32:57–74. [PubMed] [Google Scholar]
- Simonsen B, Bryan FL, Christian JHB, Roberts TA, Tompkin RB, Silliker JH. Prevention and control of foodborne salmonellosis through applicaiton of Hazard Analysis Critical Control Point (HACCP) Int J Food Microbiol. 1987;4:227–247. doi: 10.1016/0168-1605(87)90040-7. [DOI] [Google Scholar]
- Thal E, Rutqvist L, Holmqvist H. Salmonella isolated from animals in Sweden during the years 1949 to 1956. Nord Vet Med. 1957;9:822–830. [Google Scholar]
- Thorberg BM, Engvall A. Incidence of Salmonella in five Swedish slaughterhouses. J Food Prot. 2001;64:542–545. doi: 10.4315/0362-028x-64.4.542. [DOI] [PubMed] [Google Scholar]
- Thorns CJ. Bacterial food-borne zoonoses. Rev Sci Tech. 2000;19:226–239. doi: 10.20506/rst.19.1.1219. [DOI] [PubMed] [Google Scholar]
- Ward LR, Sa DEJDH, Rowe B. A phage-typing scheme for Salmonella enteritidis. Epidem Inf. 1987;99:291–294. doi: 10.1017/s0950268800067765. [DOI] [PMC free article] [PubMed] [Google Scholar]


