Abstract
Biological signals for transforming growth factor β (TGF-β) are transduced through transmembrane serine/threonine kinase receptors that signal to a family of intracellular mediators known as Smads. Smad2 and Smad4 are important for transcriptional and antiproliferative responses to TGF-β, and their inactivation in human cancers indicates that they are tumor suppressors. A missense mutation at a conserved arginine residue in the amino-terminal MH1 domain of both Smad2 and Smad4 has been identified in tumors from patients with colorectal and pancreatic cancers, respectively. However, the mechanism whereby this mutation interferes with Smad activity is uncertain. Here we show that these mutations do not disrupt activation of Smads, including receptor-mediated phosphorylation of Smad2, Smad2/Smad4 heteromeric complex formation, and Smad nuclear translocation. In contrast, we demonstrate that the mutant Smads are degraded rapidly in comparison with their wild-type counterparts. We show that this decrease in Smad protein stability occurs through induction of Smad ubiquitination by pathways involving the UbcH5 family of ubiquitin ligases. These studies thus reveal a mechanism for tumorigenesis whereby genetic defects in Smads induce their degradation through the ubiquitin-mediated pathway.
Transforming growth factor β (TGF-β), the prototypic member of a superfamily of structurally related factors, has a potent, antiproliferative effect on normal epithelial cells (1–3). Because carcinomas often escape this growth inhibitory effect, it is thought that loss of sensitivity to TGF-β may be an important contributing factor in the development of tumors (4, 5).
Biological signals for TGF-β are transduced through heteromeric complexes of two transmembrane serine/threonine kinase receptors (1–3). These receptors act in concert to activate signaling in a mechanism whereby TβRII recruits and then transphosphorylates TβRI. Signals then are propagated to a family of intracellular molecules known as Smads (reviewed in refs. 1–3 and 6). Smads can be subdivided into three classes based on their functional properties, the receptor-regulated Smads (Smad1, 2, 3, 5, and 8), the common Smads (Smad4 and 4β), and the antagonistic Smads (Smad6 and 7). Although each Smad has a distinct function, all are composed of conserved amino- and carboxyl-terminal domains known as MH1 and MH2, respectively. Studies of the TGF-β-signaling pathway have shown that upon activation of the TGF-β type I receptor, Smad2 and/or Smad3 transiently associate with the receptor and are directly phosphorylated by the receptor kinase (1–3). The phosphorylated Smad then forms a heteromeric complex with Smad4, and this complex translocates from cytoplasm into nucleus. By interacting with DNA-binding proteins, Smad complexes then positively or negatively regulate the transcription of target genes (1, 6).
Inactivating mutations in both Smad2 and Smad4 have been found in various human cancers including colorectal, lung, and pancreatic carcinomas (refs. 7–9; reviewed in ref. 5). In addition, Smad4 displays germ-line mutations in juvenile polyposis, a disease in which gastrointestinal malignancies often develop (10). In further support of the role of Smads as tumor suppressors, it has been observed that Smad4/APC double-mutant heterozygote mice develop tumors where single heterozygotes do not (11) and that Smad3 null mice also can develop tumors (12).
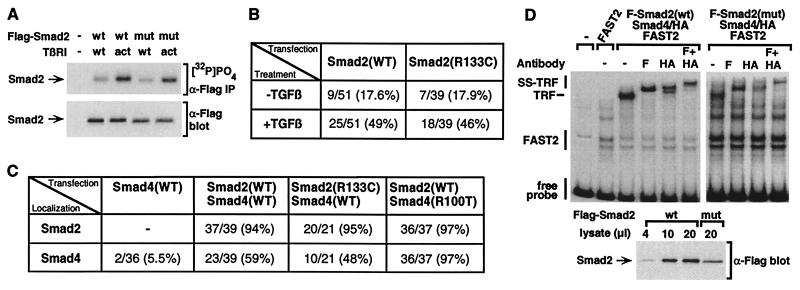
The majority of tumor-derived mutations in Smad2 and Smad4 cluster in the carboxyl-terminal MH2 domain (5), and some of these have been shown to disrupt TGF-β signaling by blocking receptor-dependent phosphorylation or by preventing heteromeric interactions between Smads (7, 13). We demonstrated previously that Smad2 harboring an arginine-to-cysteine mutation at position 133 in the amino-terminal MH1 domain (Fig. 1A) is associated with colorectal cancer (7), and mutation of this conserved arginine in Smad4 (arginine 100) also has been described in pancreatic tumors (14). To understand how these mutations might contribute to the development of cancer, we have analyzed their effects on Smad function in the TGF-β signal-transduction pathway.
Figure 1.
Loss of TGF-β-dependent transcriptional activation by MH1 domain mutants of Smad2 and Smad4. (A) Location of the MH1-domain arginine mutations in Smad2 and Smad4. (B and C) HepG2 cells, transfected with A3-Lux reporter alone or with FAST1 and wild-type (wt) or mutant (m) Smad2 or Smad4, were treated with or without TGF-β. Luciferase activity is expressed as the mean ± SD of triplicates from representative experiments.
Methods
Transcriptional Activation, Biochemical, and Electrophoretic Mobility-Shift Assays (EMSAs).
Human Smad4(R100T)/HA was subcloned into pCMV5 from pCS2-Smad4(R100T)/HA (provided by J. Massagué, Memorial Sloan–Kettering Cancer Center, New York), and all other mammalian expression constructs have been described previously (7, 15, 16). For transcriptional activation assays, HepG2 cells were transiently transfected by using calcium phosphate–DNA precipitation as described previously (17). The next day cells were treated overnight with 100 pM TGF-β and luciferase activity in cell lysates were normalized to β-galactosidase activity. For immunoprecipitations, lysates from COS-1 cells transiently transfected with DEAE-dextran were immunoprecipitated with anti-Flag M2 mAb (IBI; Eastman Kodak) and immunoblotted with anti-HA (12CA5). For Smad2 phosphorylation assays, transfected COS-1 cells were labeled with 1 mCi/ml [32P]phosphate (3,000 Ci/mmol; Amersham Pharmacia) and phosphate incorporation was visualized by immunoprecipitation and autoradiography (15). For immunofluorescence microscopy, Smad localization in transiently transfected HepG2 cells was determined as described previously (17). EMSAs were conducted as described previously (16). For transient transfec tions with wild-type or mutant Smads, equal amounts of DNA were transfected unless otherwise indicated and total DNA amounts were kept constant by the addition of pCMV5 empty vector.
Smad Ubiquitination and Steady-State Protein Level Assays.
293T cells were transiently transfected with combinations of Flag-tagged Smads, HA-tagged ubiquitin (kindly provided by D. Bohmann, European Molecular Biology Laboratory), and the wild-type His-tagged E2 ligases UbcH5a, UbcH5b, or UbcH5c (18) (kindly provided by K. Iwai, Kyoto University, Kyoto) by using calcium phosphate-mediated transfection. Lysates were immunoprecipitated with anti-Flag M2 mAb and immunoblotted with anti-HA (12CA5) mAbs. For the two-step boiling procedure, anti-Flag immunoprecipitates were boiled in 10% SDS, diluted in lysis buffer, and reprecipitated with anti-Flag antibodies before anti-HA immunoblotting. For Smad stability assays, transiently transfected COS-1 cells were labeled with 200 μCi/ml [35S]methionine (Trans [35S]-label; ICN) and then chased in methionine-containing medium in the presence or absence of 30 μM lactacystin (obtained from E. J. Corey, Harvard University). The intensity of the bands was quantitated by densitometry. To determine the effect of catalytically inactive E2s on Smad steady-state protein levels, COS-1 cells were transiently transfected with mutant Smads together with Cys-to-Ala mutant E2s (18) (provided by K. Iwai).
Results
Transcriptional Activation of a TGF-β-Responsive Promoter Is Decreased in Smad Mutants.
Mutation of a conserved arginine in the MH1 domain of Smad2 and Smad4 (Fig. 1A) has been identified in human cancers (7, 14). Because Smad2 and Smad4 are critical effectors of TGF-β signaling, we determined whether the MH1 domain mutants are capable of mediating TGF-β signals. For this, we used the TGF-β-responsive cell line, HepG2 and the A3-Lux reporter, which contains the activin-responsive element of the Mix.2 gene located upstream of a luciferase reporter gene (19, 20). As described previously (19–21), cotransfection of the winged-helix/forkhead transcription factor FAST1 with A3-Lux yielded a TGF-β-dependent induction of luciferase activity that was enhanced in cells coexpressing Smad2 and Smad4 (Fig. 1 B and C). This Smad-dependent enhancement of TGF-β signaling was not observed in cells cotransfected with equivalent amounts of Smad2(R133C) or Smad4(R100T) (Fig. 1C). A similar loss of activity by these MH1 domain mutants on 3TP-Lux, the PAI-1 promoter, and on a reporter containing a concatemerized Smad4-binding site has been described previously (22–24). Together, these data have led to the suggestion that the mutant Smad proteins are not functional.
The Smad MH1 Domain Mutants Are Phosphorylated, Form Heteromeric Complexes, and Translocate to the Nucleus.
Once phosphorylated by the type I receptor, Smad2 associates with Smad4 and this complex translocates to the nucleus (1–3). Thus, we investigated whether the MH1 domain mutants disrupted Smad function at any of these steps. We first determined whether the mutants could form heteromeric complexes in transfected COS-1 cells. As described previously, immunoprecipitation of Smad2 followed by immunoblotting for associated Smad4 revealed a low basal level of interaction between wild-type Smad2 and Smad4 that was enhanced significantly in the presence of a constitutively active TGF-β type I receptor (Fig. 2). Although initial examination of immunoprecipitates containing Smad4(R100T) appeared to indicate that complex formation was disrupted (Fig. 2A), immunoblotting of total cell lysates revealed a significant decrease in the steady-state level of Smad4 mutant protein relative to the wild type (Fig. 2A). A decrease in Smad2(R133C) protein also was observed (data not shown). To minimize this difference, we transfected higher amounts of mutant Smad plasmid DNA to obtain steady-state protein levels of mutants closer to that of wild type. Under these conditions, the MH1 domain mutants formed heteromeric complexes with their wild-type counterparts (Fig. 2 B and C). These results contrast previous findings indicating that heteromeric interactions are abrogated in Smads harboring MH1 domain mutations (22). However, in this previous study, steady-state protein levels in transfected cells were not examined, suggesting that the observed loss of interaction likely was due to reduced levels of mutant proteins.
Figure 2.
Mutation of a conserved arginine in the MH1 domain does not interfere with Smad2/Smad4 heteromeric interaction. COS-1 cells were transfected with combinations of empty vector (−), Flag-tagged wild-type (wt) or mutant (mut) Smad2 (1:2 DNA ratio) or Smad4 (1:1 DNA ratio in A; 1:4 DNA ratio in B), and wild-type (wt) or activated (act) TGF-β type I receptor (TβRI). Cell lysates were subjected to immunoprecipitation with α-Flag antibody and Smad2-associated Smad4 was visualized by α-HA immunoblotting (α-Flag IP, α-HA blot). Total protein levels were determined by immunoblotting of total cell lysates (α-Flag or α-HA blot).
To determine whether TGF-β-dependent phosphorylation of Smad2 was disrupted in Smad2 (R133C), transfected COS cells were labeled with [32P]phosphate and receptor-mediated phosphorylation of Smad2 was examined (Fig. 3A). As described previously (15), Smad2 phosphorylation was enhanced dramatically in the presence of the activated type I receptor and a similar level of phosphorylation of the MH1 domain mutant of Smad2 was observed.
Figure 3.
Effect of the MH1 domain mutation on Smad2 phosphorylation, Smad nuclear translocation, and formation of a transcriptional activation complex. (A) Smad2(R133C) is phosphorylated by the TGF-β receptor. COS-1 cells transfected with wild-type (wt) or mutant (mut) Smad2 (1:2 DNA ratio) were labeled with [32P]PO4, and Smad2 was purified by immunoprecipitation with α-Flag antibody and analyzed by autoradiography ([32P]PO4, α-Flag IP). Total Smad2 protein levels were determined by immunoblotting of total cell lysates (α-Flag blot). (B and C) HepG2 cells were transfected with wild-type or mutant Flag-Smad2 (B) or with FAST2 together with wild-type or mutant Flag-Smad2 and Smad4-HA (C). Cells were incubated with (B and C) or without (B) TGF-β for 1 h, and the subcellular localization of expressed proteins was determined by immunofluorescence microscopy. The percentage of cells in which Smad2 (B and C) or Smad4 (C) displays an exclusively nuclear localization is shown. Because Smad2 recruits Smad4 into the nucleus, the decreased level of Smad4(R100T) protein allows the nuclear accumulation of the entire mutant Smad4 pool, thereby yielding an apparent increase in nuclear accumulation of Smad4(R100T) in comparison with the wild type. (D) EMSA. COS-1 cells were transfected with constitutively active TGF-β type I receptor, FAST2, Smad4, and either wild-type or mutant Smad (1:2 DNA ratio). Equivalent amounts of nuclear extracts were incubated with a 32P-labeled 121-bp gsc promoter probe, and protein/DNA complexes were separated by nondenaturing electrophoresis. For supershifting, α-Flag (F) or α-HA (HA) antibodies were added. Total Smad2 protein levels were determined by immunoblotting of aliquots of nuclear extracts (α-Flag blot).
We next examined the nuclear accumulation of Smad2 by immunofluorescence microscopy in HepG2 cells transfected with either wild-type or mutant Flag-tagged Smad2. As described previously, in the absence of TGF-β, Smad2 was localized throughout the cell and TGF-β treatment led to its nuclear accumulation. We observed that the mutant Smad2 accumulated in the nucleus in a manner that was indistinguishable from that of the wild type (Fig. 3B). Because the nuclear accumulation of Smad4 appears to occur through cooperation with Smad2 (21, 25), we next examined Smad4 localization in cells cotransfected with Smad2. In addition, we coexpressed the nuclear target, FAST2, which promotes quantitative nuclear accumulation of Smad2 (25). Under these conditions, mutant Smad2 efficiently recruited wild-type Smad4 into the nucleus and the wild-type Smad2 promoted the nuclear accumulation of the mutant Smad4 (Fig. 3C). Because movement of Smad4 into the nucleus is thought to depend on its association with Smad2, these data also support the notion that MH1 domain mutants do not interfere with heteromeric complex formation.
Efficient transcriptional activation of FAST-target promoters requires the formation of a TGF-β/activin response factor complex (TRF) in which FAST2 and Smad4 bind to adjacent sites on the DNA while Smad2 mediates recruitment of Smad4 to FAST by associating with both proteins (16, 19, 21, 26). We assessed TRF formation by EMSAs on a fragment of the goosecoid (gsc) promoter (16). In cells expressing FAST2, Smad4, and mutant Smad2, we observed formation of a TRF. Supershift analysis confirmed the presence of mutant Smad2 in the complex (Fig. 3D). The level of TRF formation was less than that observed for complexes containing wild-type Smad2; however, this reduction is likely due to the decreased steady-state level of mutant Smad2 protein that leads to a concomitant loss of cooperative interactions between Smad complexes and FAST (Fig. 3D). In a similar analysis of mutant Smad4, we failed to detect the TRF (data not shown). However, analysis of mutant Smad4 protein by immunoblotting revealed that we were unable to achieve the minimum levels of Smad4 required for TRF formation with wild-type protein (data not shown). Of note, similar to earlier studies using different DNA fragments (23, 24, 27), binding of bacterially expressed Smad4 to the gsc promoter was lost in the mutant Smad4 (data not shown). Because we were unable to conduct EMSAs in mammalian cells, the effect of this loss of DNA binding on TRF formation is unclear. Together, the observations indicate that although the transcriptional activation of TGF-β-inducible promoters is abrogated in the MH1 domain mutants of Smad2 and Smad4, this block is not due to a lack of receptor I-mediated phosphorylation, a prevention of Smad2 and Smad4 heteromerization, or a lack of Smad nuclear accumulation.
The MH1 Domain Mutation in Smads Causes an Increased Rate of Protein Degradation.
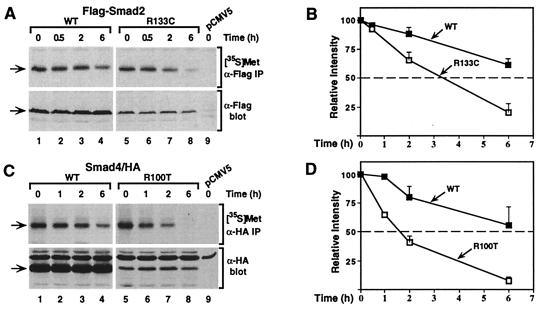
The decreased levels of the Smad mutant proteins (Fig. 2) suggested that the MH1 domain mutation leads to altered turnover of Smad proteins. To investigate this, transfected COS-1 cells were pulsed with [35S]methionine and then chased in the presence of excess unlabeled methionine. The level of newly synthesized protein in both wild-type and mutant Smad transfectants was indistinguishable; however, with increasing incubation times, the disappearance of newly synthesized mutant protein was accelerated when compared with the wild-type protein (Fig. 4 A and C). The half-life of mutant Smad2 was decreased from about 8 to 3 h, and for Smad4, the mutant exhibited a reduction in half-life from approximately 6 to 1.5 h (Fig. 4 B and D). For both Smads, the steady-state levels of wild-type and mutant proteins remained constant throughout the time course of the experiment (Fig. 4). Thus, Smad2 and Smad4 harboring an arginine mutation in the MH1 domain are translated at a similar rates but are degraded more rapidly compared with their wild-type counterparts.
Figure 4.
The mutation in Smad2 and Smad4 causes an increased rate of protein degradation. COS-1 cells transfected with wild-type (WT) or mutant (R133C) Flag-Smad2 (A) and wild-type (WT) or mutant (R100T) Smad4-HA (C) were labeled with [35S]methionine and then incubated in unlabeled culture medium for the indicated times. Cell lysates were subjected to immunoprecipitation, and labeled protein was visualized by autoradiography ([35S]Met). Aliquots of cell lysates were immunoblotted to determine the steady state of protein levels (α-Flag or α-HA blot). (B and D) The relative levels of Smad proteins were quantitated by laser densitometry and are expressed as the mean ± SD of three separate experiments, with the exception of the 0.5-h (B) and 1-h (D) time points, which represent single points.
The MH1 Domain Mutants of Smads Are Degraded Through the Ubiquitin–Proteasome Pathway.
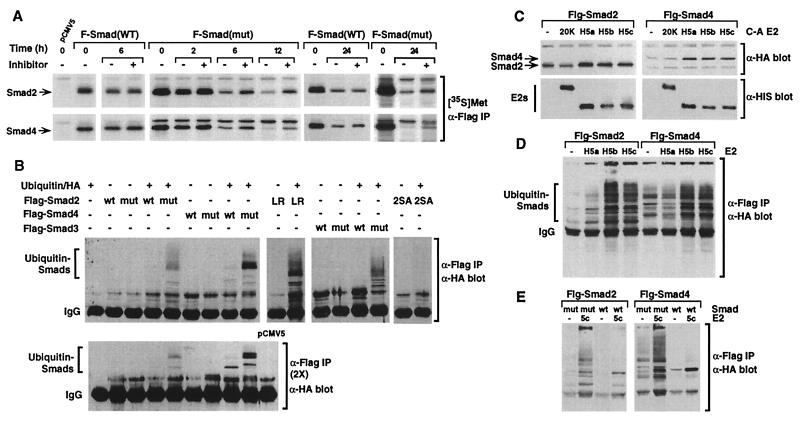
To determine whether the mutant Smad proteins were targeted for degradation through the ubiquitin–proteasome pathway (28), we examined the effect on protein stability of lactacystin, a highly specific inhibitor of the 26S proteasome (29). Lactacystin treatment of COS-1 cells transfected with Smad2(R133C) or Smad4(R100T) yielded a decrease in the rate of protein degradation of newly synthesized proteins in comparison with untreated cells at each time point (Fig. 5A). In contrast, inhibitor treatment of wild-type Smad2 or Smad4 had no effect on protein levels (Fig. 5A). Similar effects were observed with LLnL, a less specific proteasome inhibitor (data not shown).
Figure 5.
The mutant Smads are degraded through the ubiquitin–proteasome pathway. (A) COS-1 cells transfected with Flag-tagged wild-type (WT) or mutant (mut) Smads were labeled with [35S]methionine for 30 min and then incubated in unlabeled culture medium with or without lactacystin for various times. Cell lysates were immunoprecipitated with α-Flag antibody, and [35S]methionine-labeled Smads were visualized by autoradiography ([35S]Met, α-Flag IP). (B) 293T cells transfected with Flag-tagged Smads and HA-tagged ubiquitin were immunoprecipitated with α-Flag antibody and ubiquitin-conjugated Smads were detected by immunoblotting with α-HA antibody (α-Flag IP, α-HA blot). Immunoprecipitates were boiled in SDS and reprecipitated with α-Flag antibodies before α-HA immunoblotting [α-Flag IP (2×), α-HA blot]. (C) COS-1 cells transfected with Flag-tagged Smads with or without the indicated E2 dominant-negative ligases (E2 C-A) were lysed, and steady-state Smad and E2 ligase protein levels were determined by immunoblotting with α-Flag or α-His antibodies. (D and E) 293T cells transfected with Flag-tagged Smads, HA-tagged ubiquitin, and wild-type E2s were immunoprecipitated with α-Flag antibody, and ubiquitin-conjugated Smads were detected by immunoblotting with α-HA antibody (α-Flag IP, α-HA blot).
To determine whether ubiquitinated forms of the mutant Smads could be detected, 293T cells were transfected with wild-type or mutant Smads with or without HA-tagged ubiquitin. Conjugated ubiquitin was detected by Smad immunoprecipitation followed by anti-HA immunoblotting. In cells expressing mutant Smads along with HA-ubiquitin, slowly migrating bands, indicative of multiubiquitinated Smad conjugates, were observed (Fig. 5B). These bands were confirmed to represent ubiquitin-conjugated Smads by boiling the anti-Flag immunoprecipitates and reprecipitating before immunoblotting (Fig. 5B). Although no cancer-associated mutations have been identified in Smad3, substitution of threonine for the conserved MH1 domain arginine similarly yielded a highly ubiquitinated form of the mutant protein (Fig. 5B). We previously identified a mutation in the MH2 domain of Smad2 (L440R) that resulted in a dramatic reduction of steady-state protein levels (7). Similar to the MH1 domain mutants, Smad2 (L440R) yielded a multiubiquitinated form of the Smad. In contrast, mutations in the carboxyl-terminal phosphorylation sites of Smad2 (2SA), which prevents activation by the TGF-β receptor, did not yield poly-ubiquitinated Smad2. In the case of wild-type Smads, little or no Smad-ubiquitin adducts were detected. Thus, together with the proteasome inhibitor data, these findings demonstrate that the ubiquitin–proteasome pathway is important for the rapid degradation of mutant Smad2, Smad3, and Smad4.
To identify the ubiquitinating enzymes involved in degradation of the mutant Smads, we examined the effect of coexpressing catalytically inactive versions of E2s on the steady-state levels of mutant Smad2 and Smad4. These mutant E2s harbor an active-site Cys-to-Ala mutation and yield an inactive ligase that acts in a dominant-negative manner (18). We observed that the mutant versions of UbcH5a, UbcH5b, and UbcH5c caused an increase in the steady-state levels of mutant Smad2 and Smad4, whereas the ligase, E2–20K, did not (Fig. 5C). Consistent with the higher rate of degradation of mutant Smad4 compared with mutant Smad2, expression of the catalytically inactive E2s yielded a greater increase in mutant Smad4 steady-state proteins levels. To confirm the involvement of these E2s in Smad degradation, we examined the effect of coexpressing wild-type E2s on ubiquitination of the mutant Smads. Consistent with our studies using dominant-negative versions of E2s, we observed that UbcH5b and UbcH5c and, to a lesser extent, UbcH5a significantly enhanced the levels of multiubiquitinated Smad proteins (Fig. 5D). To compare ubiquitination of wild-type and mutant Smads, we focused on UbcH5c, which shares more than 90% identity with UbcH5a and UbcH5b. Coexpression of UbcH5c did not result in the generation of higher-order ubiquitin conjugates on wild-type Smads, in contrast to the abundant higher-molecular-weight species formed with mutant Smads (Fig. 5E). However, UbcH5c did lead to some ubiquitination of Smad2 that was restricted to shorter ubiquitin chains. In the case of Smad4, we detected no UbcH5c-dependent ubiquitination of Smad4, despite a strong enhancement in multiubiquitinated species of mutant Smad4. Together, our data indicate that UbcH5a, H5b, and H5c are preferentially involved in ubiquitin-mediated proteolysis of mutant Smads.
Increased Expression of Smad2 (R133C) but Not Smad4 (R100T) Restores TGF-β Signaling.
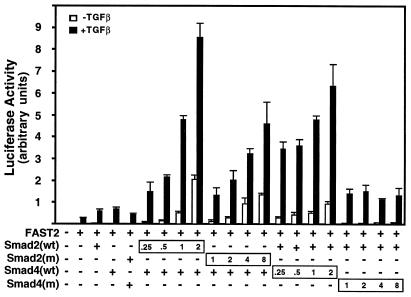
Our data suggest that the primary defect in MH1 domain mutants is to induce targeting of Smads for ubiquitin-mediated proteolysis. Thus, we examined whether elevating the level of Smad2 mutant protein might restore Smad-dependent activation of a TGF-β target gene. HepG2 cells were transfected with the FAST-dependent gsc-Lux reporter (16), FAST2, and increasing amounts of wild-type Smad2 or Smad4. Under these conditions, Smad2/Smad4 expression strongly enhanced TGF-β-dependent induction of the promoter (Fig. 6). At the lower level of transfected plasmid, Smad2(R133C) had no effect; however, at higher levels, the mutant Smad2 enhanced activation of the promoter close to levels obtained with wild-type Smad2. Increasing amounts of Smad4(R100T) failed to restore transcriptional activation of the promoter. This is consistent with our inability to detect a TRF and may be due to the more severe effect of this mutation on Smad4 stability or, alternatively, is the result of the inability of the mutant to bind DNA. Thus, elevating Smad2(R133C) is able to restore TGF-β-dependent activation of a Smad target promoter.
Figure 6.
Transcriptional activation is restored by increasing amounts of Smad2(R133C) but not Smad4(R100T). HepG2 cells were transfected with the gsc-Lux reporter alone or together with increasing amounts of wild-type (wt) or mutant versions of Smad2 or Smad4 with (+) or without (−) FAST2, as in Fig. 1. The relative levels of transfected Smad DNA constructs are indicated.
Discussion
Escape from the antiproliferative effects of TGF-β often is associated with the progression of tumorigenesis in human cancers (4). Here, we have examined the mechanism of disruption of TGF-β signaling in Smad2 and Smad4 that harbors alterations in a conserved arginine residue in the MH1 domain. We observed that the majority of Smad functions are preserved but that this mutation induces targeting of Smads for degradation through the ubiquitin–proteasome pathway. We postulate that this decrease in protein level contributes significantly to a loss in cellular responsiveness to TGF-β. Of note, bacterially expressed Smad4 harboring this mutation cannot bind DNA (this study and refs. 23, 24, and 27), suggesting the possibility that in the case of Smad4, this defect also might contribute to a loss of TGF-β signaling. It was proposed previously that mutation of the MH1 domain arginines increases potential autoinhibitory interactions between the MH1 and MH2 domains of Smads and that this causes a block in TGF-β-dependent signaling (22). Our observations that Smad steady-state levels are decreased provide an alternative mechanism for inactivation of the Smad pathway. Because cellular responses are highly sensitive to the level of Smad protein (30), we suggest that the MH1 mutations promote tumorigenesis by causing reductions in the steady-state levels of Smad protein that are insufficient to mediate TGF-β signaling. This pathway for inactivating Smads may not be restricted to MH1 domain mutants because another cancer-associated mutation that resides in the MH2 domain is similarly subject to ubiquitin-mediated proteolysis (this study).
Crystallographic analysis of the Smad3 MH1 domain structure has shown that the arginine residue mutated in cancers is located on a surface patch. Alteration of this residue is not predicted to cause major disruption to the MH1 domain structure but may cause local perturbations (31). Importantly, if these alterations occur they do not interfere with the majority of Smad functions. How mutation of the arginine induces recognition of the mutant Smad by the ubiquitin–proteasome machinery is currently unclear. Recently, it was shown that activation of the TGF-β-signaling pathway leads to ubiquitination and targeted degradation of activated, phosphorylated Smad2 (32). In our study, ubiquitination and degradation of the mutant Smads were observed in the absence of signaling, although, in both cases, members of the UbcH5 family of E2s appeared to be involved. Interestingly, an E3 HECT-domain ubiquitin ligase that specifically targets Smad1 for degradation also appears to function independently of activation of bone morphogenetic protein signaling (33). Thus, an interesting possibility in the context of tumorigenesis is that other genetic alterations in the ubiquitin–proteasome pathway, rather than in the Smads themselves, could cause induction of wild-type Smad degradation, with subsequent loss of TGF-β responsiveness.
The ubiquitin–proteasome pathway plays an important role in many cellular processes, and, not unexpectedly, this pathway has been implicated in several human diseases. For example, a block of p53 or β-catenin degradation leads to elevation in protein levels that are associated with several human cancers (28). In these examples, mutations act by disrupting the degradation of proteins that otherwise are regulated normally by the ubiquitin–proteasome pathway. In contrast, we have identified a mechanism whereby mutations in Smads do not interfere with most of the normal functions of the tumor suppressor protein, but rather inactivate the protein by inducing targeting to the ubiquitin–proteasome system. Thus, enhancing ubiquitination of tumor suppressor proteins can contribute to the development of human cancers.
Acknowledgments
We thank J. Massagué for human Smad4(R100T), D. Bohmann for HA-ubiquitin, K. Iwai for the E2s, C. Silvestri for the mutant Smad3, S. Abdollah for preliminary experiments, and J. Wrana and D. Rotin for discussions. This work was supported by grants to L.A. from the National Cancer Institute of Canada with funds from the Canadian Cancer Society. L.A. is a Medical Research Council of Canada Scholar, and J.X. was a recipient of an Natural Sciences and Engineering Research Council studentship and a Medical Research Council of Canada Fellowship.
Abbreviations
- TGF-β
transforming growth factor β
- TRF
TGF-β/activin response factor complex
- EMSA
electrophoretic mobility-shift assay
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Attisano L, Wrana J L. Curr Op Cell Biol. 2000;12:235–243. doi: 10.1016/s0955-0674(99)00081-2. [DOI] [PubMed] [Google Scholar]
- 2.Heldin C-H, Miyazono K, ten Dijke P. Nature (London) 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 3.Massagué J. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 4.Markowitz S D, Roberts A B. Cytokine Growth Factor Rev. 1996;7:93–102. doi: 10.1016/1359-6101(96)00001-9. [DOI] [PubMed] [Google Scholar]
- 5.Hata A, Shi Y, Massague J. Mol Med Today. 1998;6:257–262. doi: 10.1016/s1357-4310(98)01247-7. [DOI] [PubMed] [Google Scholar]
- 6.Derynck R, Zhang Y, Feng X-H. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- 7.Eppert K, Scherer S W, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui L-C, Bapat B, Gallinger S, Andrulis I, et al. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 8.Hahn S A, Schutte M, Shamsul Hoque A T M, Moskaluk C A, da Costa L T, Rozenblum E, Weinstein C L, Fischer A, Yeo C J, Hruban R H, et al. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 9.Riggins G J, Thiagalingam S, Rozenblum E, Weinstein C L, Kern S E, Hamilton S R, Willson J K V, Markowitz S D, Kinzler K W, Vogelstein B. Nat Genet. 1996;13:347–349. doi: 10.1038/ng0796-347. [DOI] [PubMed] [Google Scholar]
- 10.Howe J R, Roth S, Ringold J C, Summers R W, Jarvinen H J, Sistonen P, Tomlinson I P M, Houlston R S, Bevan S, Mitros F A, et al. Science. 1998;280:1086–1088. doi: 10.1126/science.280.5366.1086. [DOI] [PubMed] [Google Scholar]
- 11.Takaku K, Oshima M, Miyoshi H, Matsui M, Seldin M F, Taketo M M. Cell. 1998;92:645–656. doi: 10.1016/s0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Y, Richardson J A, Parada L F, Graff J M. Cell. 1998;94:703–714. doi: 10.1016/s0092-8674(00)81730-4. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y, Hata A, Lo R S, Massagué J, Pavletich N P. Nature (London) 1997;388:87–93. doi: 10.1038/40431. [DOI] [PubMed] [Google Scholar]
- 14.Schutte M, Hruban R H, Hedrick L, Cho K R, Nadasdy G M, Weinstein C L, Bova G S, Isaacs W B, Cairns P, Nawroz H, et al. Cancer Res. 1996;56:2527–2530. [PubMed] [Google Scholar]
- 15.Macías-Silva M, Abdollah S, Hoodless P A, Pirone R, Attisano L, Wrana J L. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 16.Labbé E, Silvestri C, Hoodless P A, Wrana J L, Attisano L. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- 17.Hoodless P A, Haerry T, Abdollah S, Stapleton M, O'Connor M B, Attisano L, Wrana J L. Cell. 1996;85:489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- 18.Gonen H, Bercovich B, Orian A, Carrano A, Takizawa C, Yamanaka K, Pagano M, Iwai K, Ciechanover A. J Biol Chem. 1999;274:14823–14830. doi: 10.1074/jbc.274.21.14823. [DOI] [PubMed] [Google Scholar]
- 19.Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Nature (London) 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu Y-Y, Grinnell B W, Richardson M A, Topper J N, Gimbrone M A, Jr, Wrana J L, et al. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 21.Liu F, Pouponnot C, Massagué J. Genes Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hata A, Lo R S, Wotton D, Lagna G, Massagué J. Nature (London) 1997;388:82–87. doi: 10.1038/40424. [DOI] [PubMed] [Google Scholar]
- 23.Dai J L, Turnacioglu K K, Schutte M, Sugar A Y, Kern S E. Cancer Res. 1998;58:4592–4597. [PubMed] [Google Scholar]
- 24.Song C-Z, Siok T E, Gelehrter T D. J Biol Chem. 1998;273:29287–29290. doi: 10.1074/jbc.273.45.29287. [DOI] [PubMed] [Google Scholar]
- 25.Hoodless P A, Tsukazaki T, Nishimatsu S-i, Attisano L, Wrana J L, Thomsen G H. Dev Biol. 1999;207:364–379. doi: 10.1006/dbio.1998.9168. [DOI] [PubMed] [Google Scholar]
- 26.Zhou S, Zawel L, Lengauer C, Kinzler K W, Vogelstein B. Mol Cell. 1998;2:121–127. doi: 10.1016/s1097-2765(00)80120-3. [DOI] [PubMed] [Google Scholar]
- 27.Stroschein S L, Wang W, Luo K. J Biol Chem. 1999;274:9431–9441. doi: 10.1074/jbc.274.14.9431. [DOI] [PubMed] [Google Scholar]
- 28.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 29.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 30.Shimizu K, Gurdon J B. Proc Natl Acad Sci USA. 1999;96:6791–6796. doi: 10.1073/pnas.96.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi Y, Wang Y-F, Jayaraman L, Yang H, Massagué J, Pavletich N P. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- 32.Lo R S, Massagué J. Nat Cell Biol. 1999;1:472–478. doi: 10.1038/70258. [DOI] [PubMed] [Google Scholar]
- 33.Zhu H, Kavsak P, Abdollah S, Wrana J L, Thomsen G H. Nature (London) 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]